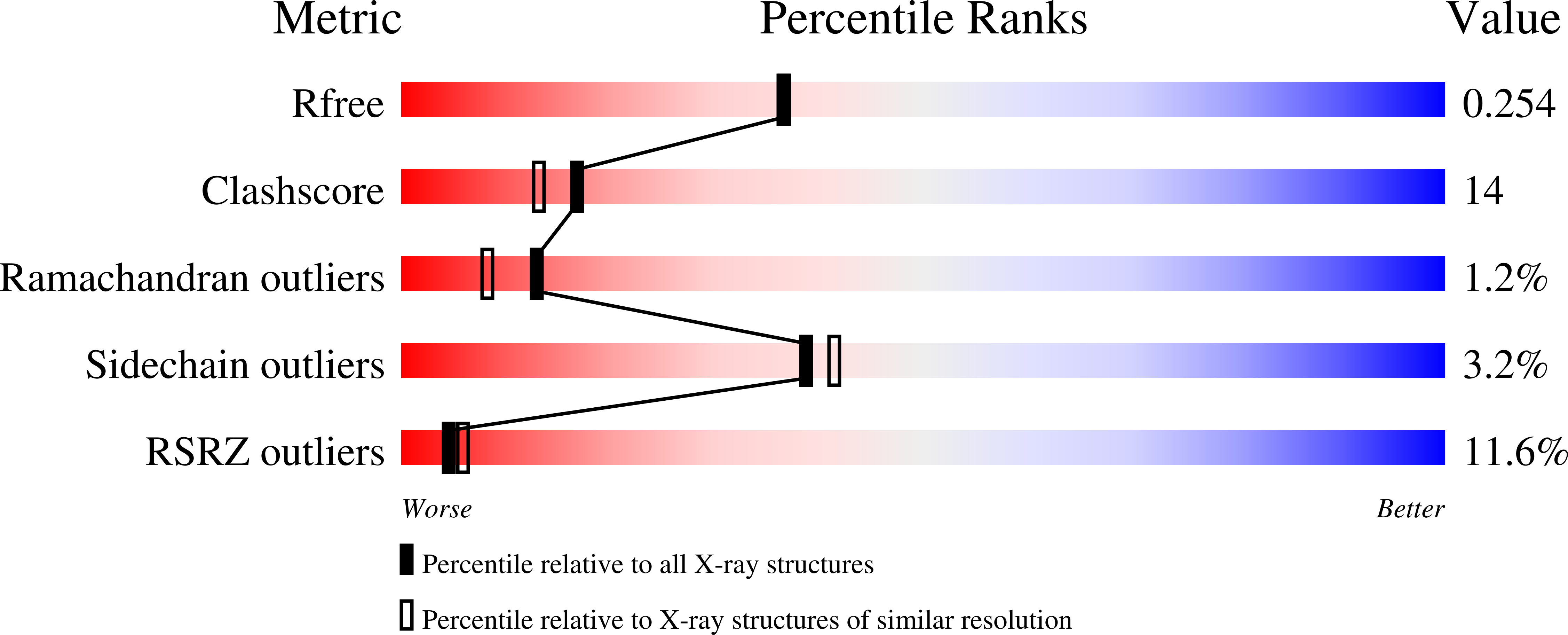
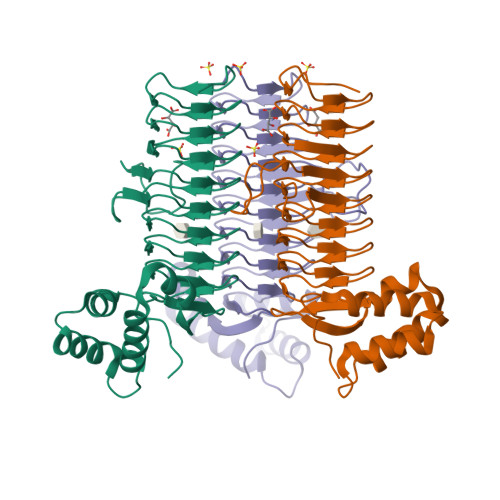
Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Lee, B.I., Suh, S.W.(2003) Proteins 53: 772-774
- PubMed: 14579368
- DOI: https://doi.org/10.1002/prot.10436
- Primary Citation of Related Structures:
1J2Z
Organizational Affiliation:
Laboratory of Structural Proteomics, School of Chemistry and Molecular Engineering, Seoul National University, Seoul, Korea.