X-ray structure of glucose/galactose receptor from Salmonella typhimurium in complex with the physiological ligand, (2R)-glyceryl-beta-D-galactopyranoside
Sooriyaarachchi, S., Ubhayasekera, W., Boos, W., Mowbray, S.L.(2009) FEBS J 276: 2116-2124
- PubMed: 19292879
- DOI: https://doi.org/10.1111/j.1742-4658.2009.06945.x
- Primary Citation of Related Structures:
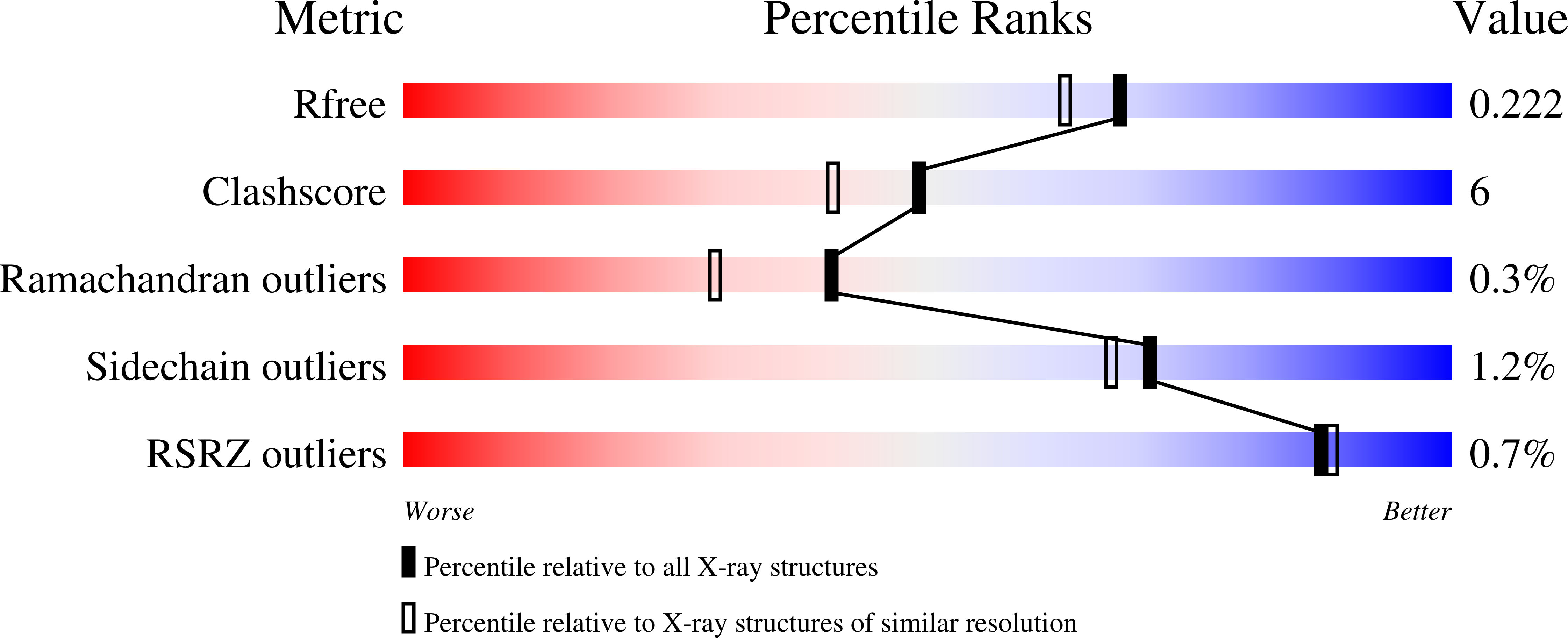
3GA5 - PubMed Abstract:
Periplasmic binding proteins are abundant in bacteria by virtue of their essential roles as high-affinity receptors in ABC transport systems and chemotaxis. One of the best studied of these receptors is the so-called glucose/galactose-binding protein. Here, we report the X-ray structure of the Salmonella typhimurium protein bound to the physiologically relevant ligand, (2R)-glyceryl-beta-D-galactopyranoside, solved by molecular replacement, and refined to 1.87 A resolution with R and R-free values of 17% and 22%. The structure identifies three amino acid residues that are diagnostic of (2R)-glyceryl-beta-D-galactopyranoside binding (Thr110, Asp154 and Gln261), as opposed to binding to the monosaccharides glucose and galactose. These three residues are conserved in essentially all available glucose/galactose-binding protein sequences, indicating that the binding of (2R)-glyceryl-beta-D-galactopyranoside is the rule rather than the exception for receptors of this type. The role of (2R)-glyceryl-beta-D-galactopyranoside in bacterial biology is discussed. Further, comparison of the available structures provides the most complete description of the conformational changes of glucose/galactose-binding protein to date. The structures follow a smooth and continuous path from the most closed structure [that bound to (2R)-glyceryl-beta-D-galactopyranoside] to the most open (an apo structure).
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Uppsala, Sweden.