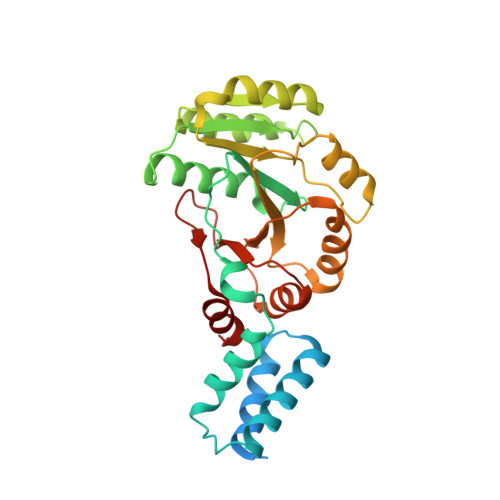
Structural basis for the molecular evolution of SRP-GTPase activation by protein.
Bange, G., Kummerer, N., Grudnik, P., Lindner, R., Petzold, G., Kressler, D., Hurt, E., Wild, K., Sinning, I.(2011) Nat Struct Mol Biol 18: 1376-1380
- PubMed: 22056770
- DOI: https://doi.org/10.1038/nsmb.2141
- Primary Citation of Related Structures:
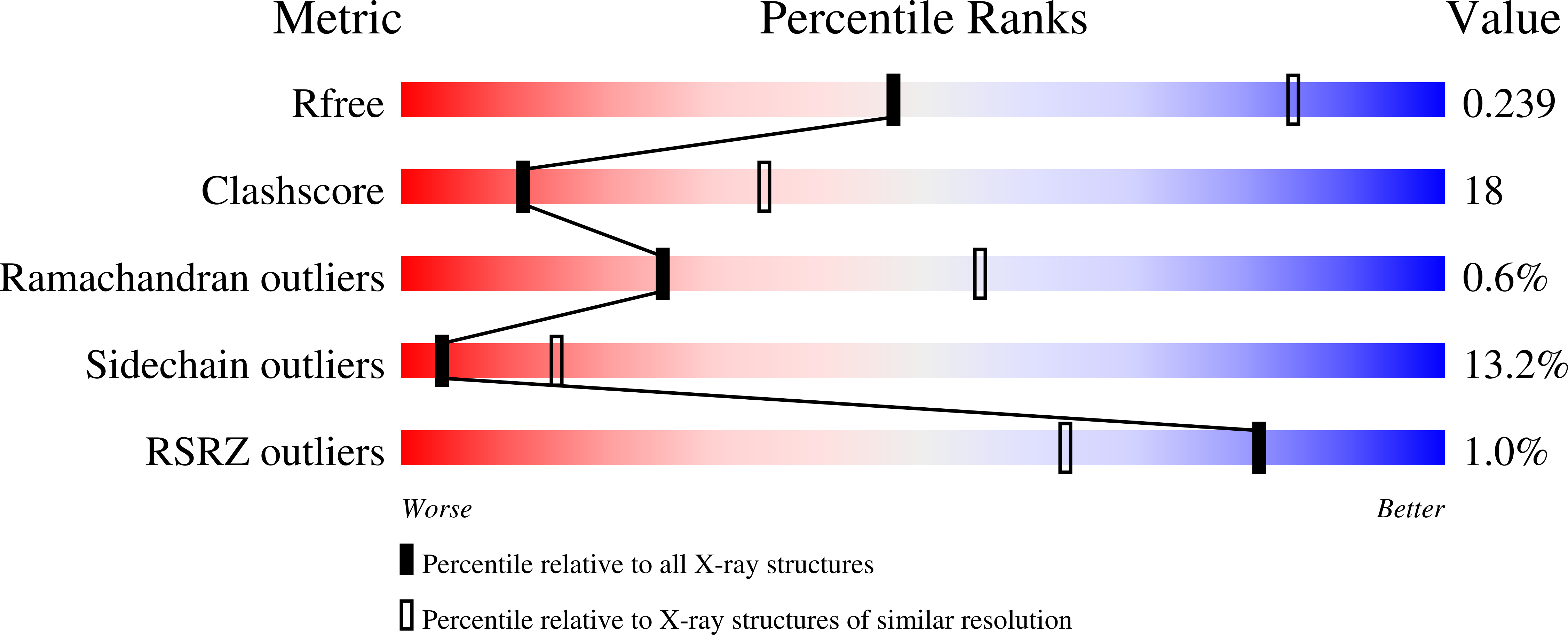
3SYN - PubMed Abstract:
Small G proteins have key roles in signal transduction pathways. They are switched from the signaling 'on' to the non-signaling 'off' state when GTPase-activating proteins (GAPs) provide a catalytic residue. The ancient signal recognition particle (SRP)-type GTPases form GTP-dependent homo- and heterodimers and deviate from the canonical switch paradigm in that no GAPs have been identified. Here we show that the YlxH protein activates the SRP-GTPase FlhF. The crystal structure of the Bacillus subtilis FlhF-effector complex revealed that the effector does not contribute a catalytic residue but positions the catalytic machinery already present in SRP-GTPases. We provide a general concept that might also apply to the RNA-driven activation of the universally conserved, co-translational protein-targeting machinery comprising the SRP-GTPases Ffh and FtsY. Our study exemplifies the evolutionary transition from RNA- to protein-driven activation in SRP-GTPases and suggests that the current view on SRP-mediated protein targeting is incomplete.
Organizational Affiliation:
Heidelberg University Biochemistry Center, Heidelberg, Germany. gert.bange@bzh.uni-heidelberg.de