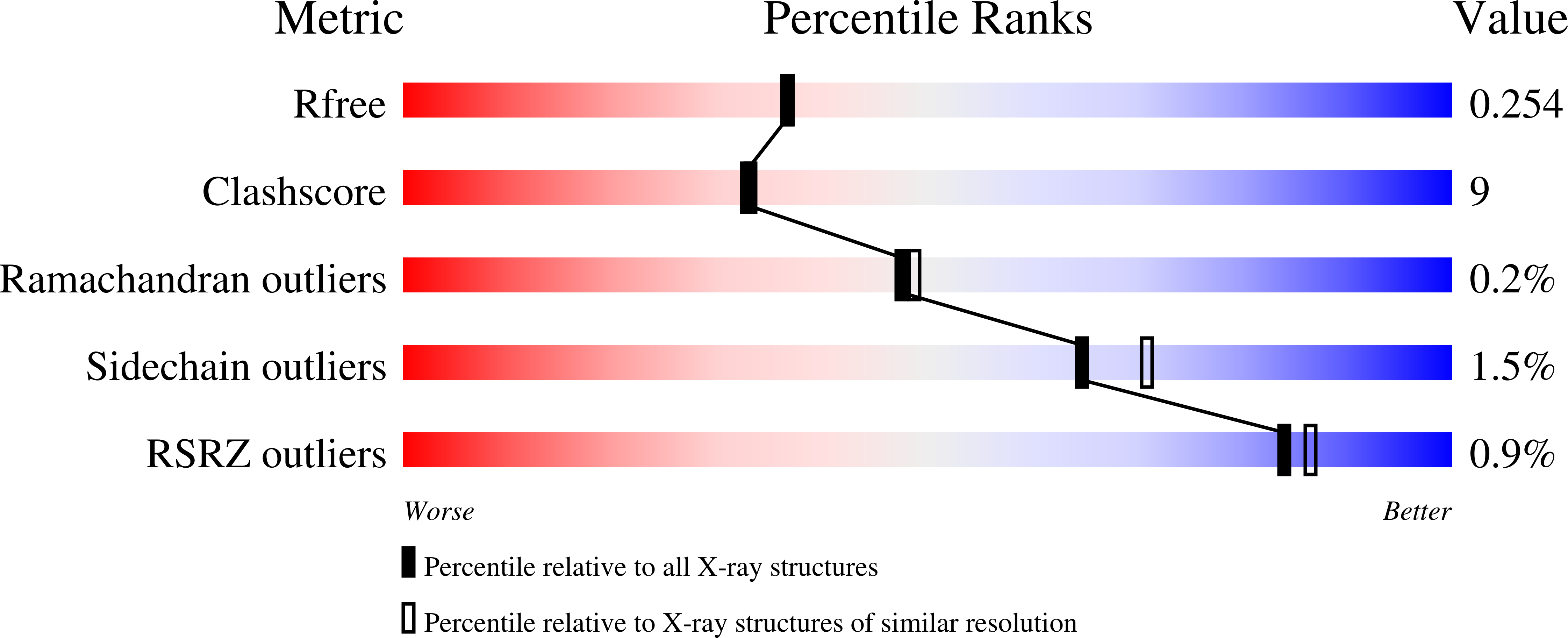
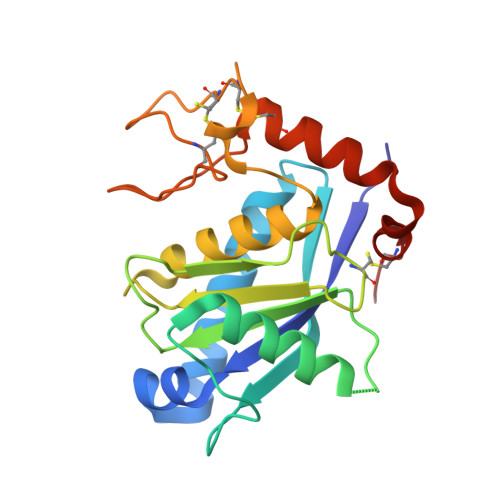
Structure of human ADAM-8 catalytic domain complexed with batimastat.
Hall, T., Shieh, H.S., Day, J.E., Caspers, N., Chrencik, J.E., Williams, J.M., Pegg, L.E., Pauley, A.M., Moon, A.F., Krahn, J.M., Fischer, D.H., Kiefer, J.R., Tomasselli, A.G., Zack, M.D.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 616-621
- PubMed: 22684055
- DOI: https://doi.org/10.1107/S1744309112015618
- Primary Citation of Related Structures:
4DD8 - PubMed Abstract:
The role of ADAM-8 in cancer and inflammatory diseases such as allergy, arthritis and asthma makes it an attractive target for drug development. Therefore, the catalytic domain of human ADAM-8 was expressed, purified and crystallized in complex with a hydroxamic acid inhibitor, batimastat. The crystal structure of the enzyme-inhibitor complex was refined to 2.1 Å resolution. ADAM-8 has an overall fold similar to those of other ADAM members, including a central five-stranded β-sheet and a catalytic Zn(2+) ion. However, unique differences within the S1' binding loop of ADAM-8 are observed which might be exploited to confer specificity and selectivity to ADAM-8 competitive inhibitors for the treatment of diseases involving this enzyme.
Organizational Affiliation:
Pfizer Inc, 700 Chesterfield Parkway West, Chesterfield, MO 63017, USA.