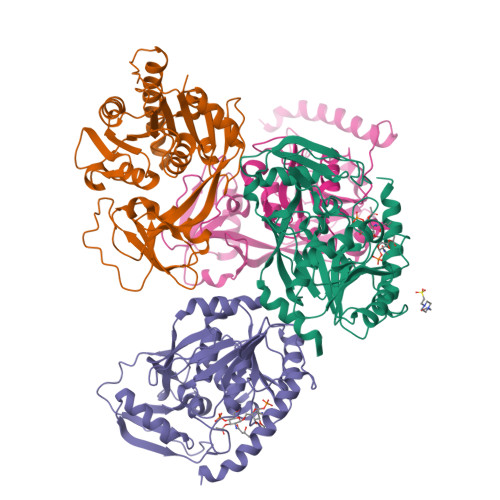
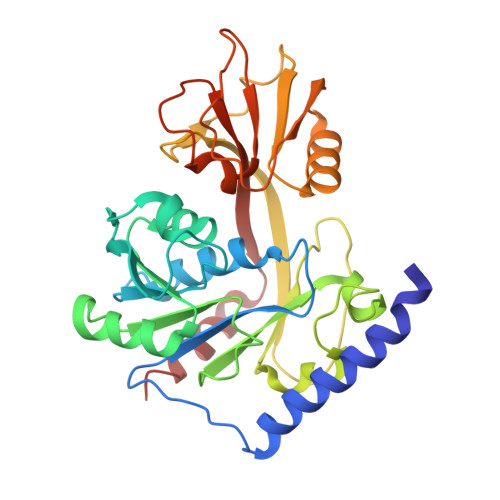
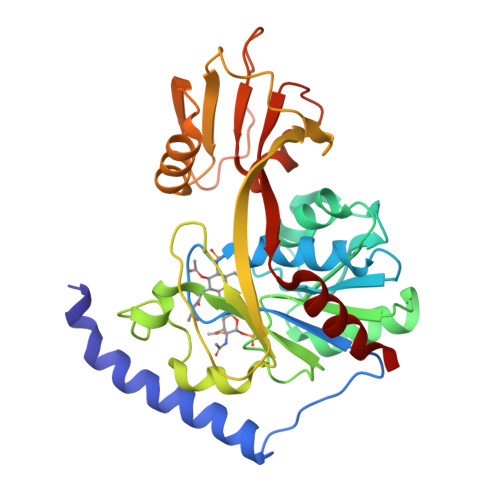
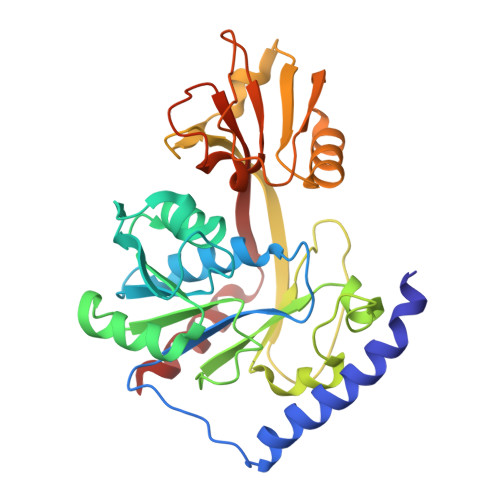
Structural Basis of Lipid Binding for the Membrane-embedded Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK.
Emptage, R.P., Tonthat, N.K., York, J.D., Schumacher, M.A., Zhou, P.(2014) J Biol Chem 289: 24059-24068
- PubMed: 25023290
- DOI: https://doi.org/10.1074/jbc.M114.589986
- Primary Citation of Related Structures:
4LKV - PubMed Abstract:
The membrane-bound tetraacyldisaccharide-1-phosphate 4'-kinase, LpxK, catalyzes the sixth step of the lipid A (Raetz) biosynthetic pathway and is a viable antibiotic target against emerging Gram-negative pathogens. We report the crystal structure of lipid IVA, the LpxK product, bound to the enzyme, providing a rare glimpse into interfacial catalysis and the surface scanning strategy by which many poorly understood lipid modification enzymes operate. Unlike the few previously structurally characterized proteins that bind lipid A or its precursors, LpxK binds almost exclusively to the glucosamine/phosphate moieties of the lipid molecule. Steady-state kinetic analysis of multiple point mutants of the lipid-binding pocket pinpoints critical residues involved in substrate binding, and characterization of N-terminal helix truncation mutants uncovers the role of this substructure as a hydrophobic membrane anchor. These studies make critical contributions to the limited knowledge surrounding membrane-bound enzymes that act upon lipid substrates and provide a structural template for designing small molecule inhibitors targeting this essential kinase.
Organizational Affiliation:
From the Department of Biochemistry, Duke University Medical Center, Durham, North Carolina 27710 and.