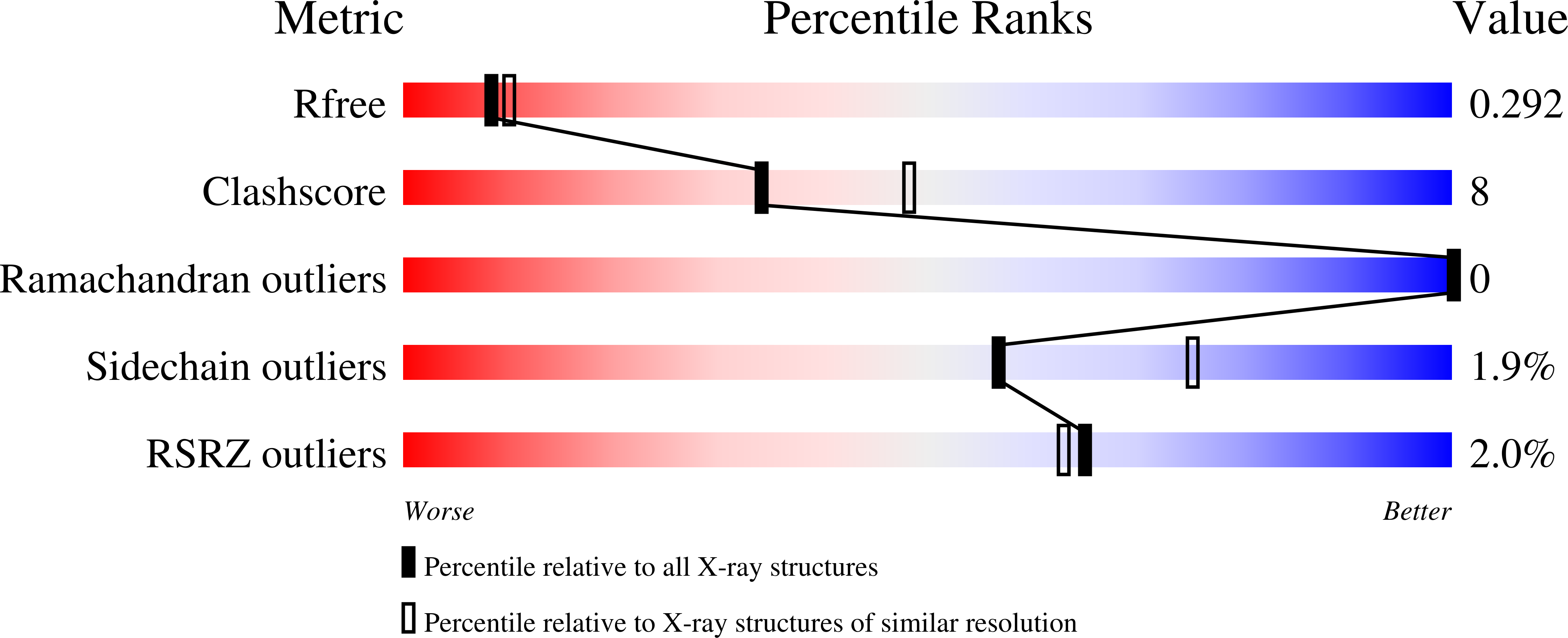
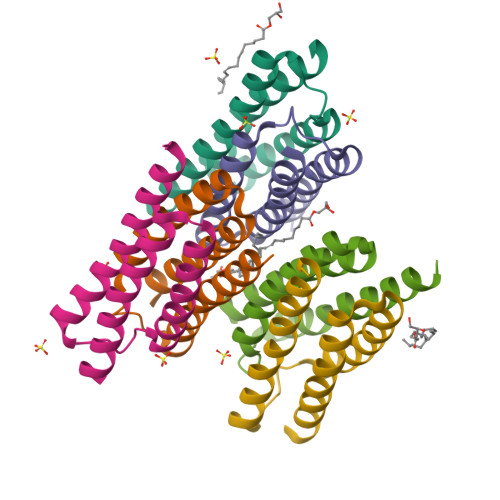
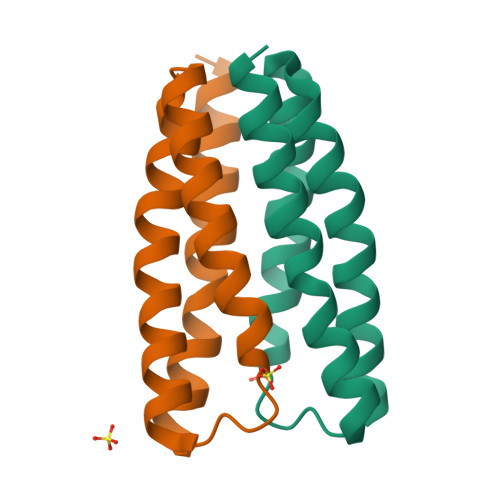
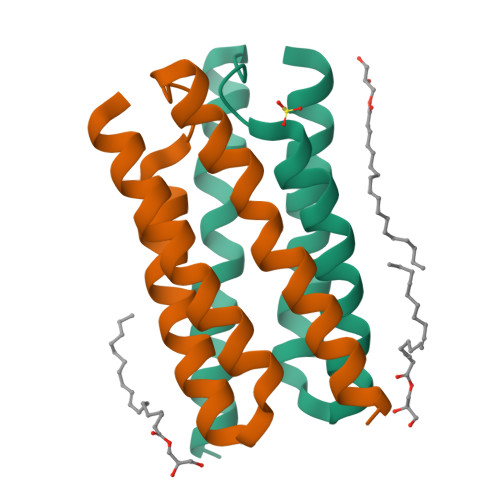
Crystal structure of a bacterial homologue of SWEET transporters.
Wang, J., Yan, C., Li, Y., Hirata, K., Yamamoto, M., Yan, N., Hu, Q.(2014) Cell Res 24: 1486-1489
- PubMed: 25378180
- DOI: https://doi.org/10.1038/cr.2014.144
- Primary Citation of Related Structures:
4RNG
Organizational Affiliation:
1] State Key Laboratory of Bio-membrane and Membrane Biotechnology, Tsinghua University, Beijing 100084, China [2] Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University, Beijing 100084, China [3] Center for Structural Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.