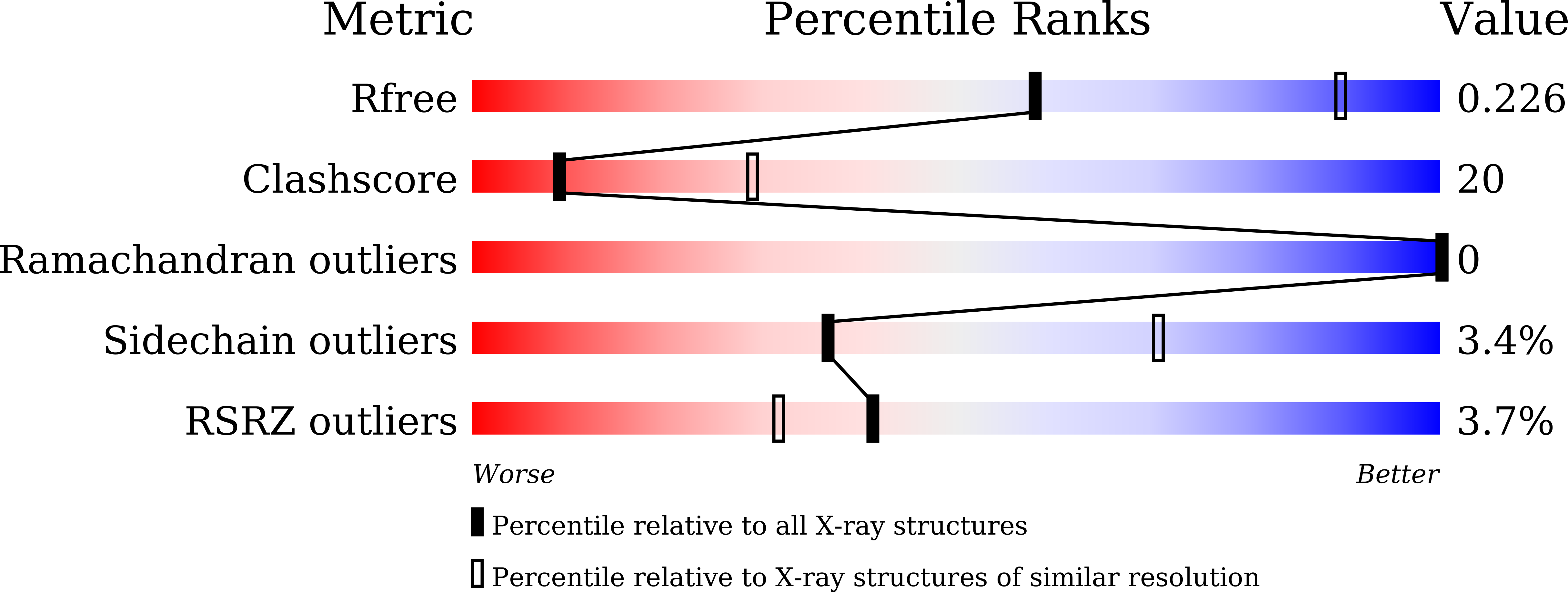
Loop L1 governs the DNA-binding specificity and order for RecA-catalyzed reactions in homologous recombination and DNA repair
Shinohara, T., Ikawa, S., Iwasaki, W., Hiraki, T., Hikima, T., Mikawa, T., Arai, N., Kamiya, N., Shibata, T.(2015) Nucleic Acids Res 43: 973-986
- PubMed: 25561575
- DOI: https://doi.org/10.1093/nar/gku1364
- Primary Citation of Related Structures:
4TWZ - PubMed Abstract:
In all organisms, RecA-family recombinases catalyze homologous joint formation in homologous genetic recombination, which is essential for genome stability and diversification. In homologous joint formation, ATP-bound RecA/Rad51-recombinases first bind single-stranded DNA at its primary site and then interact with double-stranded DNA at another site. The underlying reason and the regulatory mechanism for this conserved binding order remain unknown. A comparison of the loop L1 structures in a DNA-free RecA crystal that we originally determined and in the reported DNA-bound active RecA crystals suggested that the aspartate at position 161 in loop L1 in DNA-free RecA prevented double-stranded, but not single-stranded, DNA-binding to the primary site. This was confirmed by the effects of the Ala-replacement of Asp-161 (D161A), analyzed directly by gel-mobility shift assays and indirectly by DNA-dependent ATPase activity and SOS repressor cleavage. When RecA/Rad51-recombinases interact with double-stranded DNA before single-stranded DNA, homologous joint-formation is suppressed, likely by forming a dead-end product. We found that the D161A-replacement reduced this suppression, probably by allowing double-stranded DNA to bind preferentially and reversibly to the primary site. Thus, Asp-161 in the flexible loop L1 of wild-type RecA determines the preference for single-stranded DNA-binding to the primary site and regulates the DNA-binding order in RecA-catalyzed recombinase reactions.
Organizational Affiliation:
Cellular & Molecular Biology Unit, RIKEN, 2-1 Hirosawa, Wako-shi, Saitama 351-0198, Japan Advanced Catalysis Research Group, RIKEN Center for Sustainable Resource Science, Wako-shi, Saitama 351-0198, Japan Department of Supramolecular Biology, Graduate School of Nanobiosciences, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan.