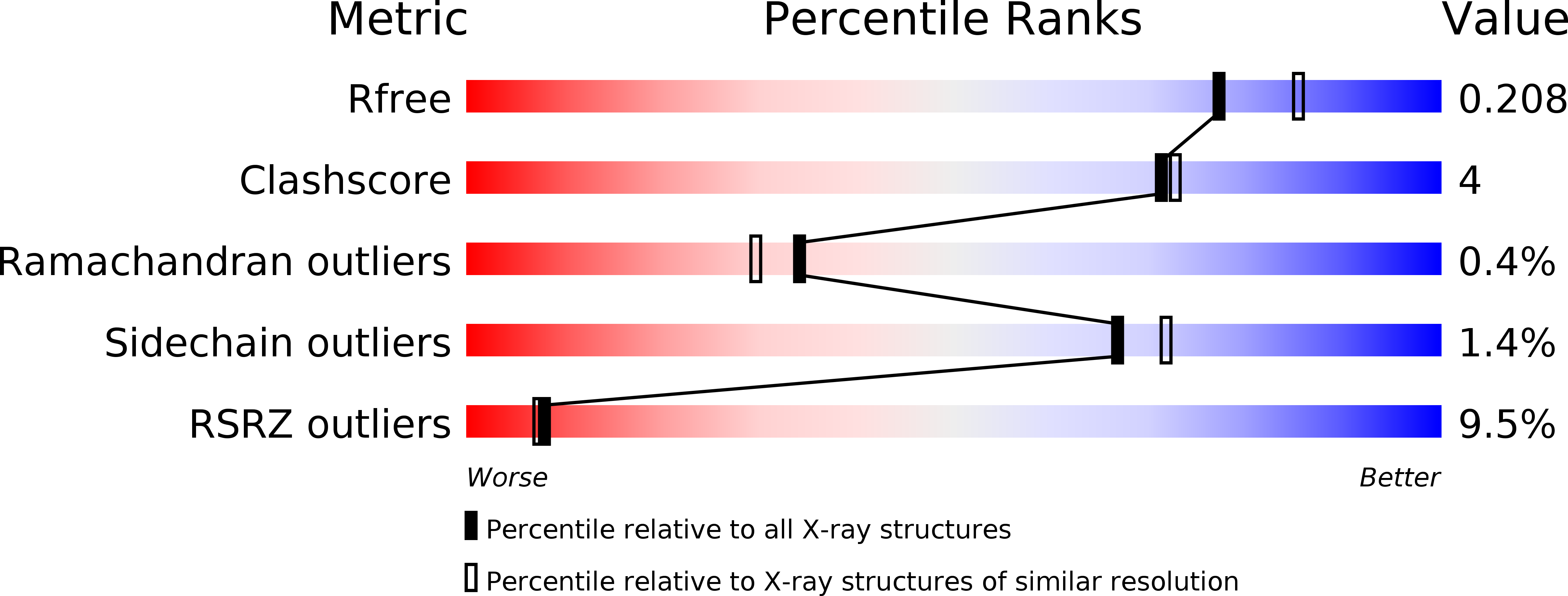
The Crystal Structure of RosB: Insights into the Reaction Mechanism of the First Member of a Family of Flavodoxin-like Enzymes.
Konjik, V., Brunle, S., Demmer, U., Vanselow, A., Sandhoff, R., Ermler, U., Mack, M.(2017) Angew Chem Int Ed Engl 56: 1146-1151
- PubMed: 27981706
- DOI: https://doi.org/10.1002/anie.201610292
- Primary Citation of Related Structures:
5MJI - PubMed Abstract:
8-demethyl-8-aminoriboflavin-5'-phosphate (AFP) synthase (RosB) catalyzes the key reaction of roseoflavin biosynthesis by forming AFP from riboflavin-5'-phosphate (RP) and glutamate via the intermediates 8-demethyl-8-formylriboflavin-5'-phosphate (OHC-RP) and 8-demethyl-8-carboxylriboflavin-5'-phosphate (HO 2 C-RP). To understand this reaction in which a methyl substituent of an aromatic ring is replaced by an amine we structurally characterized RosB in complex with OHC-RP (2.0 Å) and AFP (1.7 Å). RosB is composed of four flavodoxin-like subunits which have been upgraded with specific extensions and a unique C-terminal arm. It appears that RosB has evolved from an electron- or hydride-transferring flavoprotein to a sophisticated multi-step enzyme which uses RP as a substrate (and not as a cofactor). Structure-based active site analysis was complemented by mutational and isotope-based mass-spectrometric data to propose an enzymatic mechanism on an atomic basis.
Organizational Affiliation:
Mannheim University of Applied Sciences, Paul-Wittsack-Strasse 10, 68163, Mannheim, Germany.