Lysosomal integral membrane protein-2 as a phospholipid receptor revealed by biophysical and cellular studies.
Conrad, K.S., Cheng, T.W., Ysselstein, D., Heybrock, S., Hoth, L.R., Chrunyk, B.A., Am Ende, C.W., Krainc, D., Schwake, M., Saftig, P., Liu, S., Qiu, X., Ehlers, M.D.(2017) Nat Commun 8: 1908-1908
- PubMed: 29199275
- DOI: https://doi.org/10.1038/s41467-017-02044-8
- Primary Citation of Related Structures:
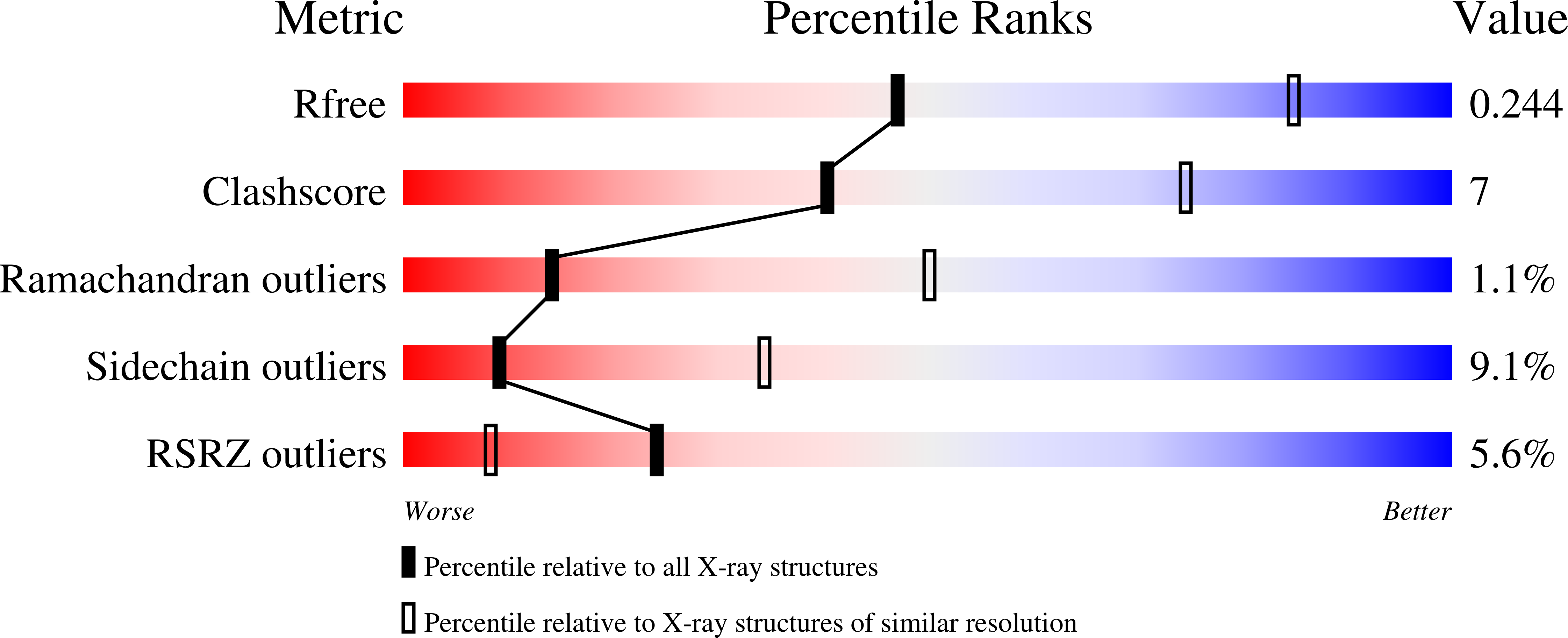
5UPH - PubMed Abstract:
Lysosomal integral membrane protein-2 (LIMP-2/SCARB2) contributes to endosomal and lysosomal function. LIMP-2 deficiency is associated with neurological abnormalities and kidney failure and, as an acid glucocerebrosidase receptor, impacts Gaucher and Parkinson's diseases. Here we report a crystal structure of a LIMP-2 luminal domain dimer with bound cholesterol and phosphatidylcholine. Binding of these lipids alters LIMP-2 from functioning as a glucocerebrosidase-binding monomer toward a dimeric state that preferentially binds anionic phosphatidylserine over neutral phosphatidylcholine. In cellular uptake experiments, LIMP-2 facilitates transport of phospholipids into murine fibroblasts, with a strong substrate preference for phosphatidylserine. Taken together, these biophysical and cellular studies define the structural basis and functional importance of a form of LIMP-2 for lipid trafficking. We propose a model whereby switching between monomeric and dimeric forms allows LIMP-2 to engage distinct binding partners, a mechanism that may be shared by SR-BI and CD36, scavenger receptor proteins highly homologous to LIMP-2.
Organizational Affiliation:
Medicinal Sciences, Pfizer Worldwide R&D, Eastern Point Road, Groton, CT, 06340, USA.