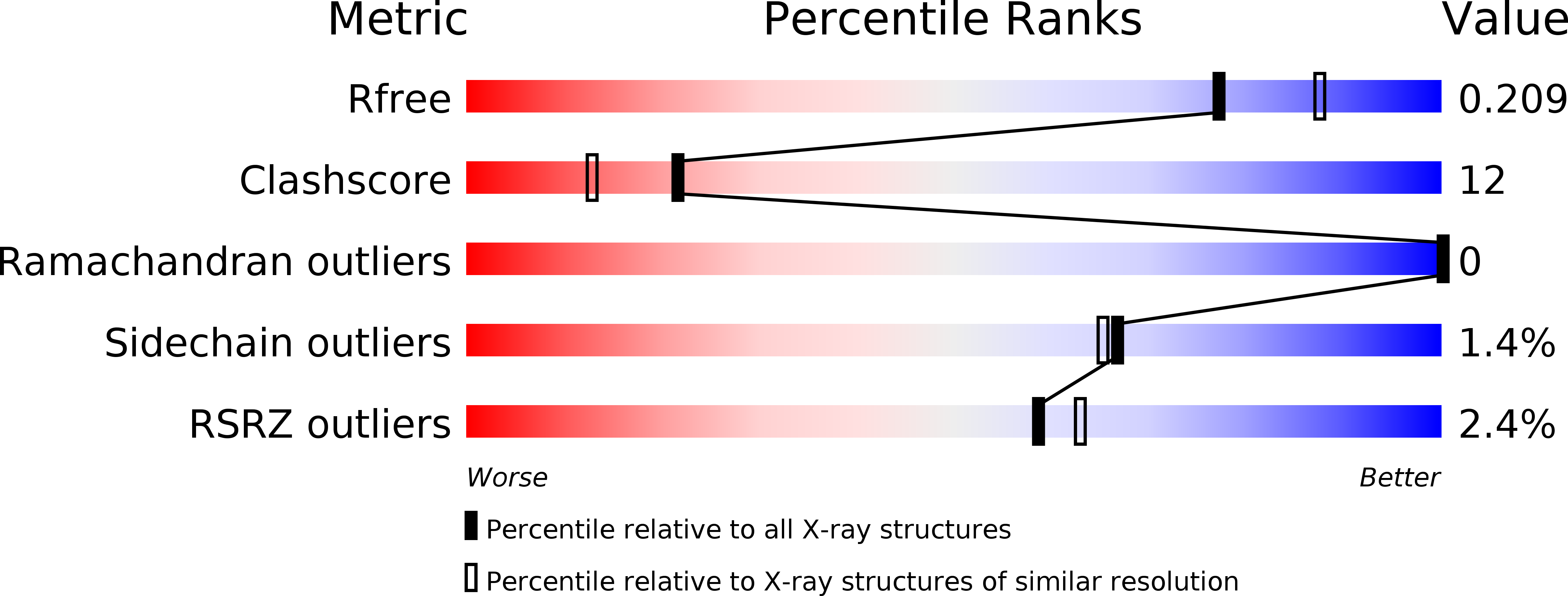
Crystal structure of l-glutamate N-acetyltransferase ArgA from Mycobacterium tuberculosis
Yang, X., Wu, L., Ran, Y., Xu, A., Zhang, B., Yang, X., Zhang, R., Rao, Z., Li, J.(2017) Biochim Biophys Acta 1865: 1800-1807
- PubMed: 28943401
- DOI: https://doi.org/10.1016/j.bbapap.2017.09.009
- Primary Citation of Related Structures:
5YGE - PubMed Abstract:
l-arginine is used as a source of both carbon and nitrogen in Mycobacterium tuberculosis (Mtb) and its biosynthesis is essential for the pathogen's survival. MtbArgA (Rv2747) catalyzes the initial step in l-arginine biosynthesis by transferring an acetyl group from acetyl coenzyme A (AcCoA) to l-glutamate. MtbArgA is a class III N-acetylglutamate synthase (NAGS) with no structural information. Here, we solved the crystal structure of MtbArgA complexed with AcCoA and l-glutamate. The overall structure adopts a classic fold of the GCN5-related N-acetyltransferase (GNAT) family, characterized by a "V"-shaped cleft and β-bulge, but uses distinct residues for the binding and reaction of AcCoA. In particular, its activity depends on dimerization to form a deep, vast pocket for l-glutamate binding. Interestingly, in the structure, l-glutamate binds at a site far away from AcCoA, implying a mechanism of separate capture and catalysis. Additionally, based on a docking model of l-glutamate at the catalytic site, a one-step sequential mechanism was proposed for enzymatic catalysis. Important sites for substrate binding and catalysis were also evaluated by site-directed mutagenesis study and activity analysis. The unique features of the MtbArgA structure will provide useful insights for inhibitor design and anti-tuberculosis drug discovery.
Organizational Affiliation:
National Center for Protein Science Shanghai, Shanghai Science Research Center, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, 333 Haike Road, Shanghai 201210, China.