A ligand binding to FXR
Yi, L., Yong, L.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bile acid receptor | 229 | Homo sapiens | Mutation(s): 0 Gene Names: NR1H4, BAR, FXR, HRR1, RIP14 | 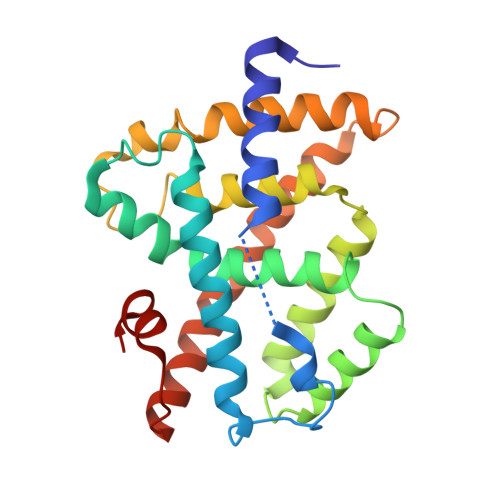 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96RI1 (Homo sapiens) Explore Q96RI1 Go to UniProtKB: Q96RI1 | |||||
PHAROS: Q96RI1 GTEx: ENSG00000012504 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96RI1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptide from Nuclear receptor coactivator 2 | 11 | Homo sapiens | Mutation(s): 0 | 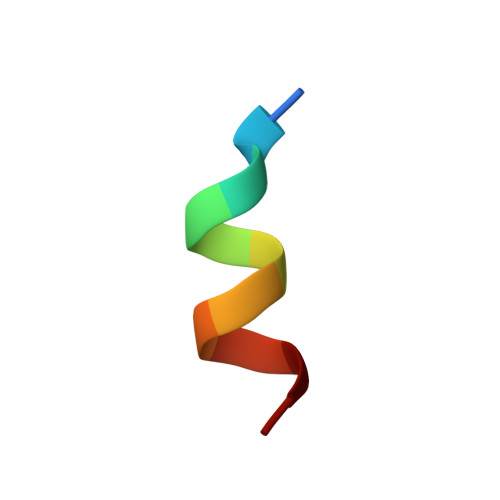 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15596 (Homo sapiens) Explore Q15596 Go to UniProtKB: Q15596 | |||||
PHAROS: Q15596 GTEx: ENSG00000140396 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15596 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 93O Query on 93O | C [auth A] | 2-methoxyethyl (2E)-3-phenylprop-2-en-1-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate C27 H26 N2 O7 HUTPFIMQINDZDT-DHZHZOJOSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 86.399 | α = 90 |
| b = 86.399 | β = 90 |
| c = 75.106 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHASER | phasing |