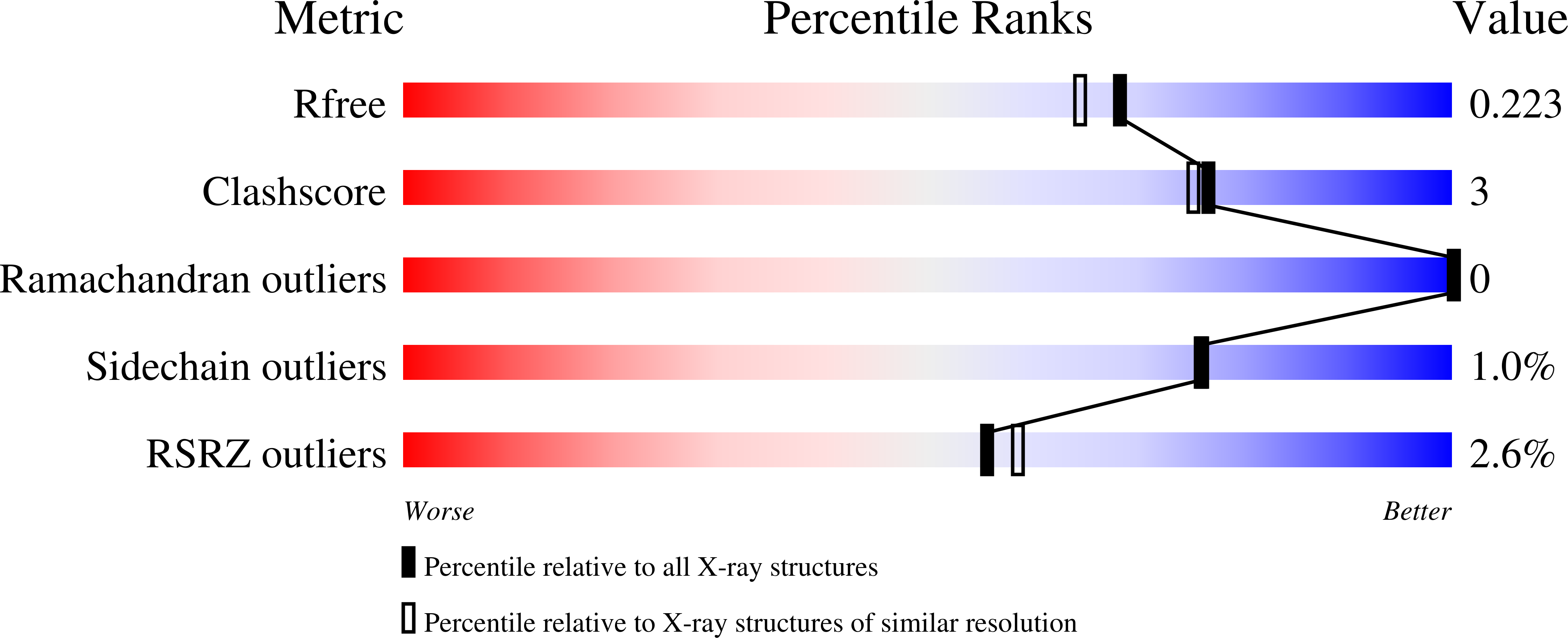
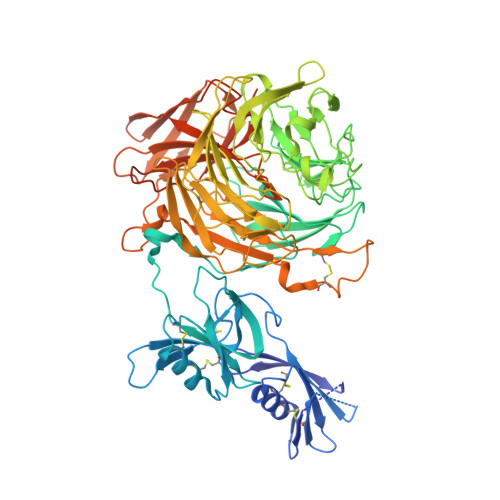
Discovery of a Fungal Copper Radical Oxidase with High Catalytic Efficiency toward 5-Hydroxymethylfurfural and Benzyl Alcohols for Bioprocessing
Mathieu, Y., Viborg, A.H., Forget, S.M., Brumer, H., Ciano, L., Offen, W.A., Blagova, E., Walton, P.H., Davies, G.J.(2020) ACS Catal