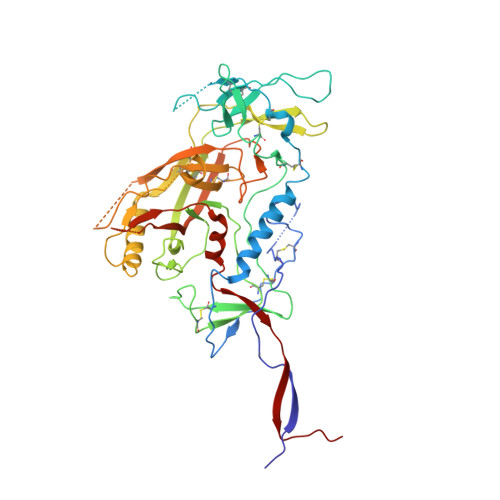
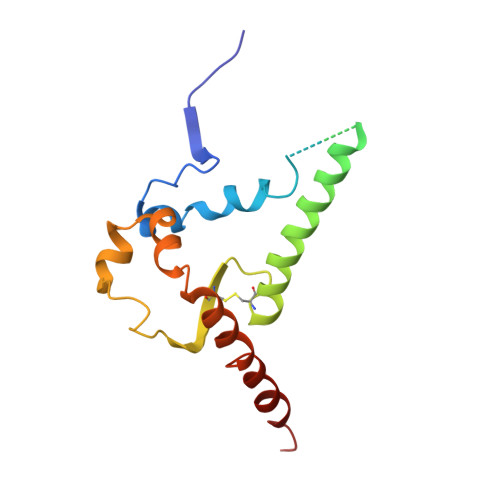
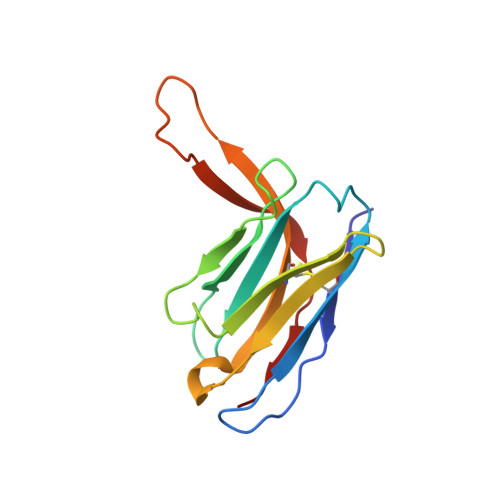
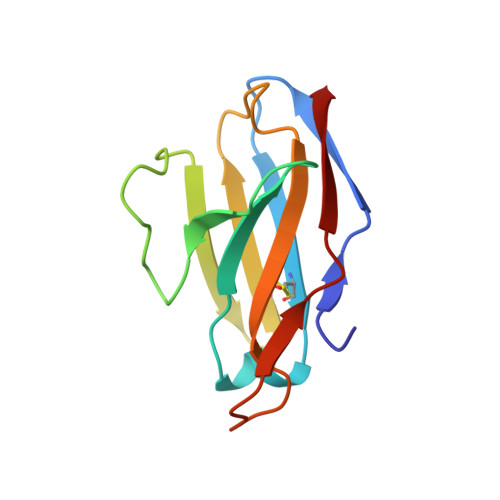
HIV-1 Envelope and MPER Antibody Structures in Lipid Assemblies.
Rantalainen, K., Berndsen, Z.T., Antanasijevic, A., Schiffner, T., Zhang, X., Lee, W.H., Torres, J.L., Zhang, L., Irimia, A., Copps, J., Zhou, K.H., Kwon, Y.D., Law, W.H., Schramm, C.A., Verardi, R., Krebs, S.J., Kwong, P.D., Doria-Rose, N.A., Wilson, I.A., Zwick, M.B., Yates 3rd, J.R., Schief, W.R., Ward, A.B.(2020) Cell Rep 31: 107583-107583
- PubMed: 32348769
- DOI: https://doi.org/10.1016/j.celrep.2020.107583
- Primary Citation of Related Structures:
6VPX - PubMed Abstract:
Structural and functional studies of HIV envelope glycoprotein (Env) as a transmembrane protein have long been complicated by challenges associated with inherent flexibility of the molecule and the membrane-embedded hydrophobic regions. Here, we present approaches for incorporating full-length, wild-type HIV-1 Env, as well as C-terminally truncated and stabilized versions, into lipid assemblies, providing a modular platform for Env structural studies by single particle electron microscopy. We reconstitute a full-length Env clone into a nanodisc, complex it with a membrane-proximal external region (MPER) targeting antibody 10E8, and structurally define the full quaternary epitope of 10E8 consisting of lipid, MPER, and ectodomain contacts. By aligning this and other Env-MPER antibody complex reconstructions with the lipid bilayer, we observe evidence of Env tilting as part of the neutralization mechanism for MPER-targeting antibodies. We also adapt the platform toward vaccine design purposes by introducing stabilizing mutations that allow purification of unliganded Env with a peptidisc scaffold.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, 92037, USA; International AIDS Vaccine Initiative Neutralizing Antibody Center, The Scripps Research Institute, La Jolla, CA 92037, USA.