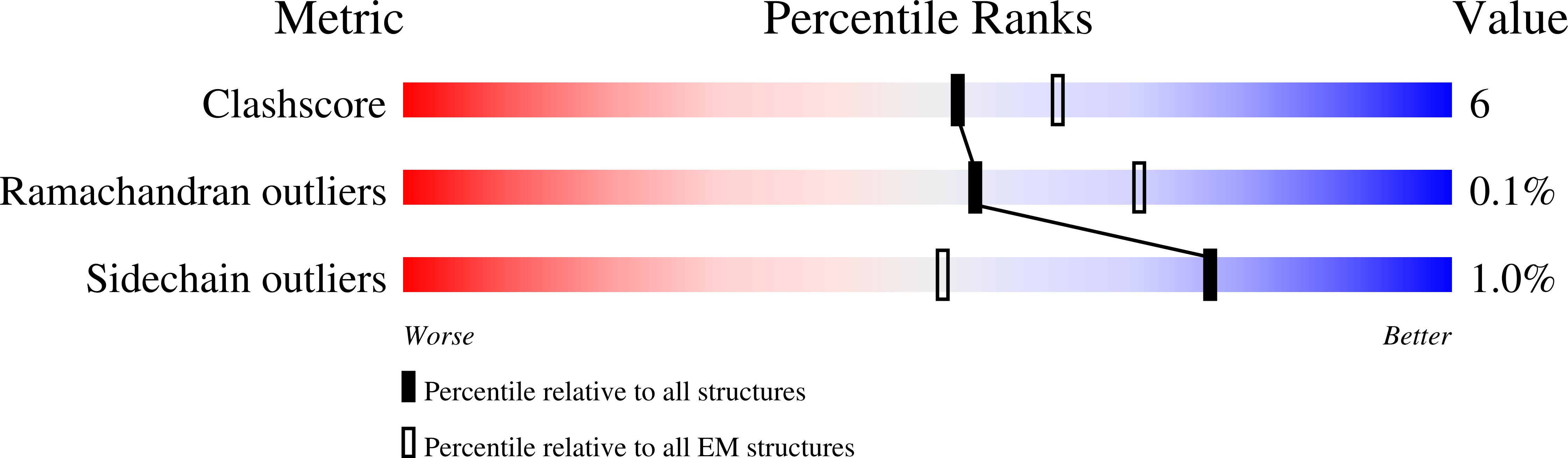Structural basis for CSPG4 as a receptor for TcdB and a therapeutic target in Clostridioides difficile infection.
Chen, P., Zeng, J., Liu, Z., Thaker, H., Wang, S., Tian, S., Zhang, J., Tao, L., Gutierrez, C.B., Xing, L., Gerhard, R., Huang, L., Dong, M., Jin, R.(2021) Nat Commun 12: 3748-3748
- PubMed: 34145250
- DOI: https://doi.org/10.1038/s41467-021-23878-3
- Primary Citation of Related Structures:
7ML7 - PubMed Abstract:
C. difficile is a major cause of antibiotic-associated gastrointestinal infections. Two C. difficile exotoxins (TcdA and TcdB) are major virulence factors associated with these infections, and chondroitin sulfate proteoglycan 4 (CSPG4) is a potential receptor for TcdB, but its pathophysiological relevance and the molecular details that govern recognition remain unknown. Here, we determine the cryo-EM structure of a TcdB-CSPG4 complex, revealing a unique binding site spatially composed of multiple discontinuous regions across TcdB. Mutations that selectively disrupt CSPG4 binding reduce TcdB toxicity in mice, while CSPG4-knockout mice show reduced damage to colonic tissues during C. difficile infections. We further show that bezlotoxumab, the only FDA approved anti-TcdB antibody, blocks CSPG4 binding via an allosteric mechanism, but it displays low neutralizing potency on many TcdB variants from epidemic hypervirulent strains due to sequence variations in its epitopes. In contrast, a CSPG4-mimicking decoy neutralizes major TcdB variants, suggesting a strategy to develop broad-spectrum therapeutics against TcdB.
Organizational Affiliation:
Department of Physiology and Biophysics, University of California, Irvine, CA, USA.