An NmrA-like enzyme-catalysed redox-mediated Diels-Alder cycloaddition with anti-selectivity.
Liu, Z., Rivera, S., Newmister, S.A., Sanders, J.N., Nie, Q., Liu, S., Zhao, F., Ferrara, J.D., Shih, H.W., Patil, S., Xu, W., Miller, M.D., Phillips, G.N., Houk, K.N., Sherman, D.H., Gao, X.(2023) Nat Chem 15: 526-534
- PubMed: 36635598
- DOI: https://doi.org/10.1038/s41557-022-01117-6
- Primary Citation of Related Structures:
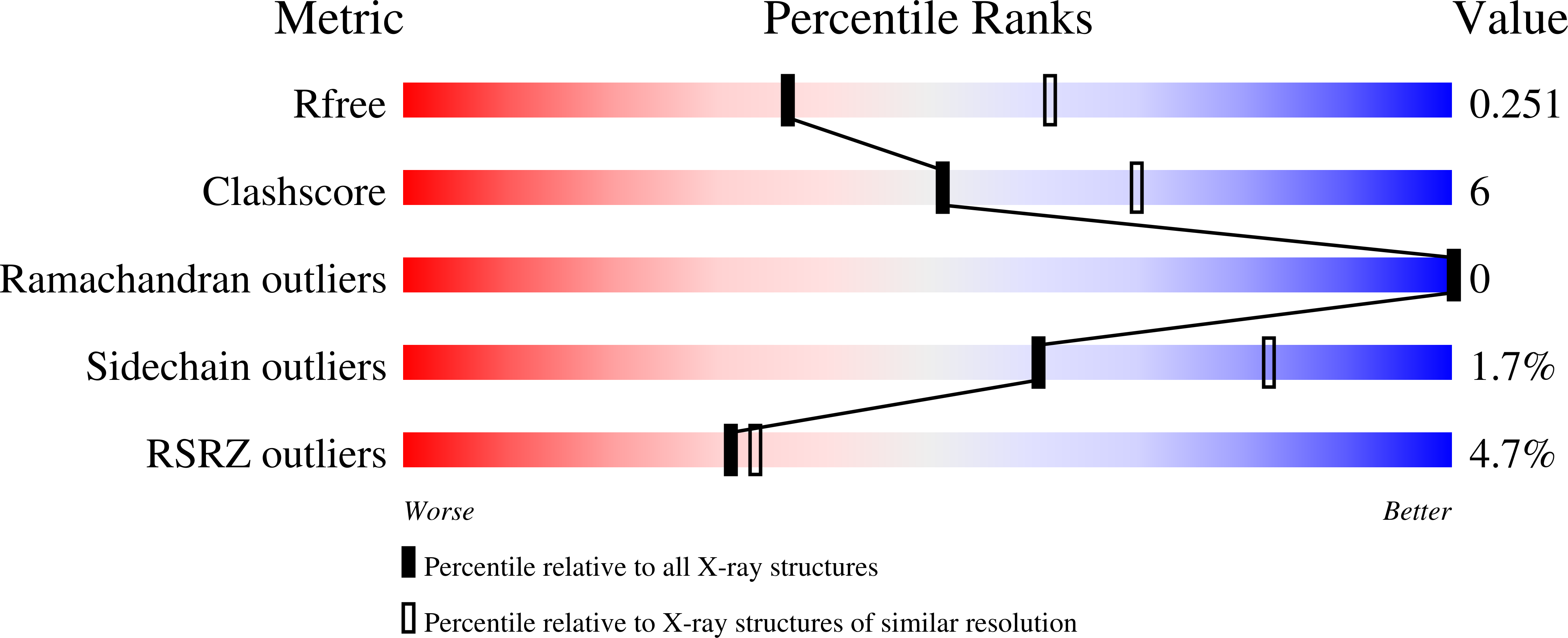
7UF8 - PubMed Abstract:
The Diels-Alder cycloaddition is one of the most powerful approaches in organic synthesis and is often used in the synthesis of important pharmaceuticals. Yet, strictly controlling the stereoselectivity of the Diels-Alder reactions is challenging, and great efforts are needed to construct complex molecules with desired chirality via organocatalysis or transition-metal strategies. Nature has evolved different types of enzymes to exquisitely control cyclization stereochemistry; however, most of the reported Diels-Alderases have been shown to only facilitate the energetically favourable diastereoselective cycloadditions. Here we report the discovery and characterization of CtdP, a member of a new class of bifunctional oxidoreductase/Diels-Alderase, which was previously annotated as an NmrA-like transcriptional regulator. We demonstrate that CtdP catalyses the inherently disfavoured cycloaddition to form the bicyclo[2.2.2]diazaoctane scaffold with a strict α-anti-selectivity. Guided by computational studies, we reveal a NADP + /NADPH-dependent redox mechanism for the CtdP-catalysed inverse electron demand Diels-Alder cycloaddition, which serves as the first example of a bifunctional Diels-Alderase that utilizes this mechanism.
Organizational Affiliation:
Department of Chemical and Biomolecular Engineering, Rice University, Houston, TX, USA.