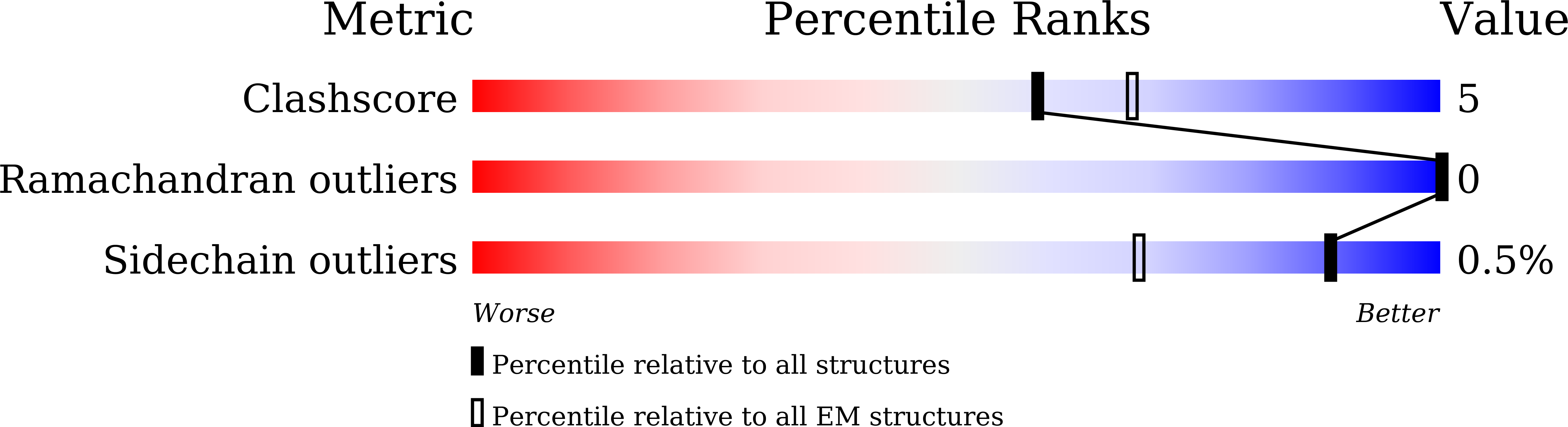Structural remodeling of ribosome associated Hsp40-Hsp70 chaperones during co-translational folding.
Chen, Y., Tsai, B., Li, N., Gao, N.(2022) Nat Commun 13: 3410-3410
- PubMed: 35701497
- DOI: https://doi.org/10.1038/s41467-022-31127-4
- Primary Citation of Related Structures:
7X34, 7X3K - PubMed Abstract:
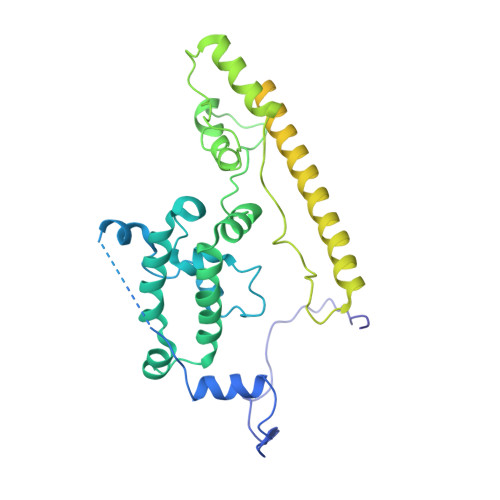
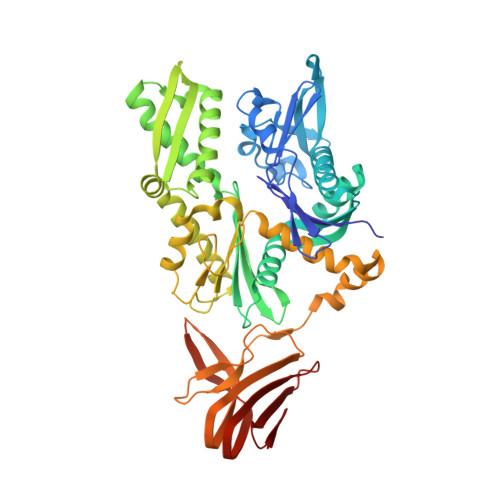
Ribosome associated complex (RAC), an obligate heterodimer of HSP40 and HSP70 (Zuo1 and Ssz1 in yeast), is conserved in eukaryotes and functions as co-chaperone for another HSP70 (Ssb1/2 in yeast) to facilitate co-translational folding of nascent polypeptides. Many mechanistic details, such as the coordination of one HSP40 with two HSP70s and the dynamic interplay between RAC-Ssb and growing nascent chains, remain unclear. Here, we report three sets of structures of RAC-containing ribosomal complexes isolated from Saccharomyces cerevisiae. Structural analyses indicate that RAC on the nascent-chain-free ribosome is in an autoinhibited conformation, and in the presence of a nascent chain at the peptide tunnel exit (PTE), RAC undergoes large-scale structural remodeling to make Zuo1 J-Domain more accessible to Ssb. Our data also suggest a role of Zuo1 in orienting Ssb-SBD proximal to the PTE for easy capture of the substrate. Altogether, in accordance with previous data, our work suggests a sequence of structural remodeling events for RAC-Ssb during co-translational folding, triggered by the binding and passage of growing nascent chain from one to another.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, School of Life Science, Tsinghua University, Beijing, 100084, China.