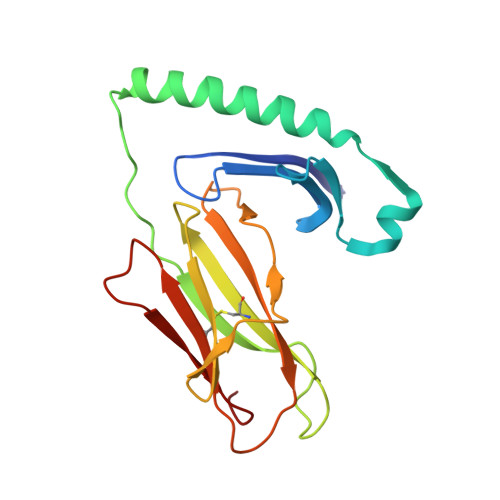
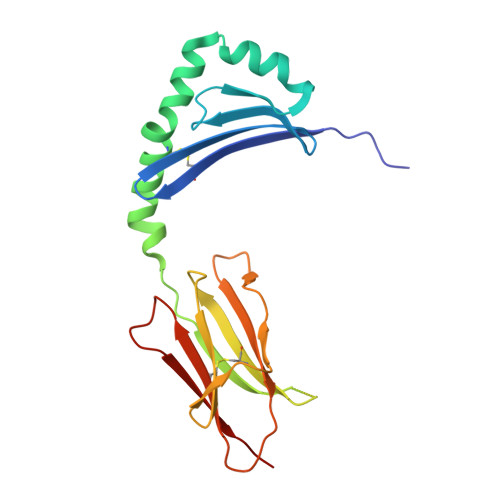
Human T cells recognize HLA-DP-bound peptides in two orientations.
Klobuch, S., Lim, J.J., van Balen, P., Kester, M.G.D., de Klerk, W., de Ru, A.H., Pothast, C.R., Jedema, I., Drijfhout, J.W., Rossjohn, J., Reid, H.H., van Veelen, P.A., Falkenburg, J.H.F., Heemskerk, M.H.M.(2022) Proc Natl Acad Sci U S A 119: e2214331119-e2214331119
- PubMed: 36442096
- DOI: https://doi.org/10.1073/pnas.2214331119
- Primary Citation of Related Structures:
7T6I - PubMed Abstract:
Human leukocyte antigen (HLA) molecules present small peptide antigens to T cells, thereby allowing them to recognize pathogen-infected and cancer cells. A central dogma over the last 50+ y is that peptide binding to HLA molecules is mediated by the docking of side chains of particular amino acids in the peptide into pockets in the HLA molecules in a conserved N- to C-terminal orientation. Whether peptides can be presented in a reversed C- to N-terminal orientation remains unclear. Here, we performed large-scale identification of peptides bound to HLA-DP molecules and observed that in addition to peptide binding in an N- to C-terminal orientation, in 9 out of 14 HLA-DP allotypes, reverse motifs are found, compatible with C- to N-terminal peptide binding. Moreover, we isolated high-avidity human cytomegalovirus (CMV)-specific HLA-DP-restricted CD4 + T cells from the memory repertoire of healthy donors and demonstrate that such T cells recognized CMV-derived peptides bound to HLA-DPB1*01:01 or *05:01 in a reverse C- to N-terminal manner. Finally, we obtained a high-resolution HLA-DPB1*01:01-CMVpp65 (142-158) peptide crystal structure, which is the molecular basis for C- to N-terminal peptide binding to HLA-DP. Our results point to unique features of HLA-DP molecules that substantially broaden the HLA class II bound peptide repertoire to combat pathogens and eliminate cancer cells.
Organizational Affiliation:
Department of Hematology, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands.