Structural and biophysical characterization of the secreted, beta-helical adhesin EtpA of Enterotoxigenic Escherichia coli.
Ntui, C.M., Fleckenstein, J.M., Schubert, W.D.(2023) PLoS One 18: e0287100-e0287100
- PubMed: 37343026
- DOI: https://doi.org/10.1371/journal.pone.0287100
- Primary Citation of Related Structures:
8CPK - PubMed Abstract:
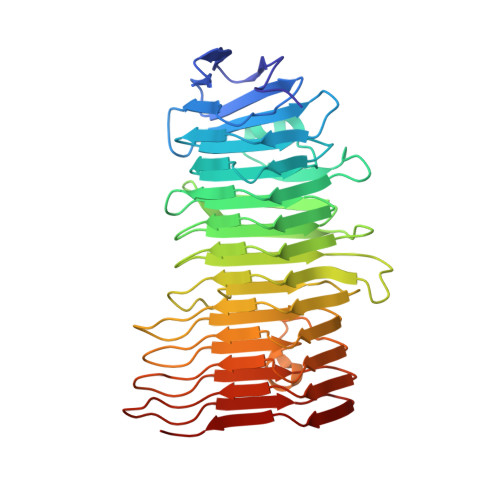
Enterotoxigenic Escherichia coli (ETEC) is a diarrhoeal pathogen associated with high morbidity and mortality especially among young children in developing countries. At present, there is no vaccine for ETEC. One candidate vaccine antigen, EtpA, is a conserved secreted adhesin that binds to the tips of flagellae to bridge ETEC to host intestinal glycans. EtpA is exported through a Gram-negative, two-partner secretion system (TPSS, type Vb) comprised of the secreted EtpA passenger (TpsA) protein and EtpB (TpsB) transporter that is integrated into the outer bacterial membrane. TpsA proteins share a conserved, N-terminal TPS domain followed by an extensive C-terminal domain with divergent sequence repeats. Two soluble, N-terminal constructs of EtpA were prepared and analysed respectively including residues 67 to 447 (EtpA67-447) and 1 to 606 (EtpA1-606). The crystal structure of EtpA67-447 solved at 1.76 Å resolution revealed a right-handed parallel β-helix with two extra-helical hairpins and an N-terminal β-strand cap. Analyses by circular dichroism spectroscopy confirmed the β-helical fold and indicated high resistance to chemical and thermal denaturation as well as rapid refolding. A theoretical AlphaFold model of full-length EtpA largely concurs with the crystal structure adding an extended β-helical C-terminal domain after an interdomain kink. We propose that robust folding of the TPS domain upon secretion provides a template to extend the N-terminal β-helix into the C-terminal domains of TpsA proteins.
Organizational Affiliation:
Department of Biochemistry, Genetics and Microbiology, University of Pretoria, Pretoria, South Africa.