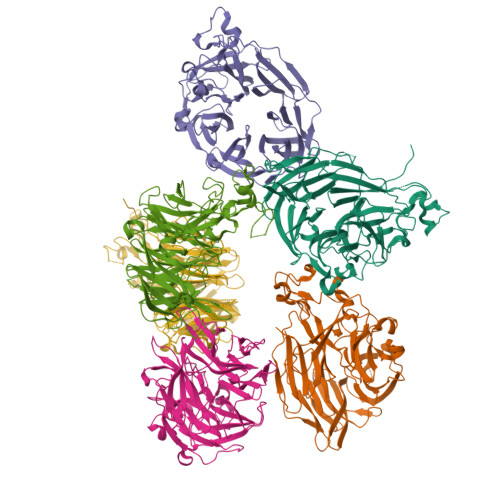Iron acquisition by a commensal bacterium modifies host nutritional immunity during Salmonella infection.
Spiga, L., Fansler, R.T., Perera, Y.R., Shealy, N.G., Munneke, M.J., David, H.E., Torres, T.P., Lemoff, A., Ran, X., Richardson, K.L., Pudlo, N., Martens, E.C., Folta-Stogniew, E., Yang, Z.J., Skaar, E.P., Byndloss, M.X., Chazin, W.J., Zhu, W.(2023) Cell Host Microbe 31: 1639-1654.e10
- PubMed: 37776864
- DOI: https://doi.org/10.1016/j.chom.2023.08.018
- Primary Citation of Related Structures:
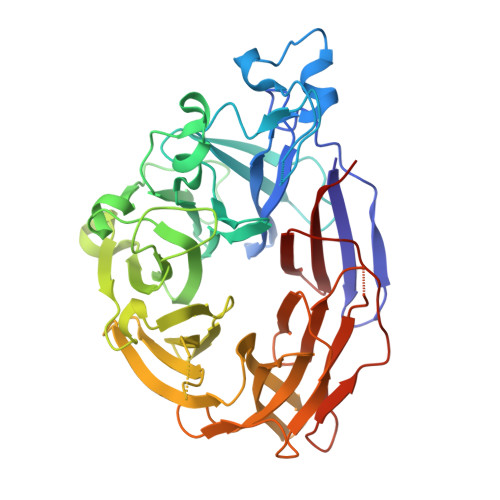
8GEX - PubMed Abstract:
During intestinal inflammation, host nutritional immunity starves microbes of essential micronutrients, such as iron. Pathogens scavenge iron using siderophores, including enterobactin; however, this strategy is counteracted by host protein lipocalin-2, which sequesters iron-laden enterobactin. Although this iron competition occurs in the presence of gut bacteria, the roles of commensals in nutritional immunity involving iron remain unexplored. Here, we report that the gut commensal Bacteroides thetaiotaomicron acquires iron and sustains its resilience in the inflamed gut by utilizing siderophores produced by other bacteria, including Salmonella, via a secreted siderophore-binding lipoprotein XusB. Notably, XusB-bound enterobactin is less accessible to host sequestration by lipocalin-2 but can be "re-acquired" by Salmonella, allowing the pathogen to evade nutritional immunity. Because the host and pathogen have been the focus of studies of nutritional immunity, this work adds commensal iron metabolism as a previously unrecognized mechanism modulating the host-pathogen interactions and nutritional immunity.
Organizational Affiliation:
Department of Pathology, Microbiology, and Immunology, Vanderbilt University Medical Center, Nashville, TN 37232, USA; Vanderbilt Institute for Infection, Immunology, and Inflammation, Vanderbilt University Medical Center, Nashville, TN 37232, USA.