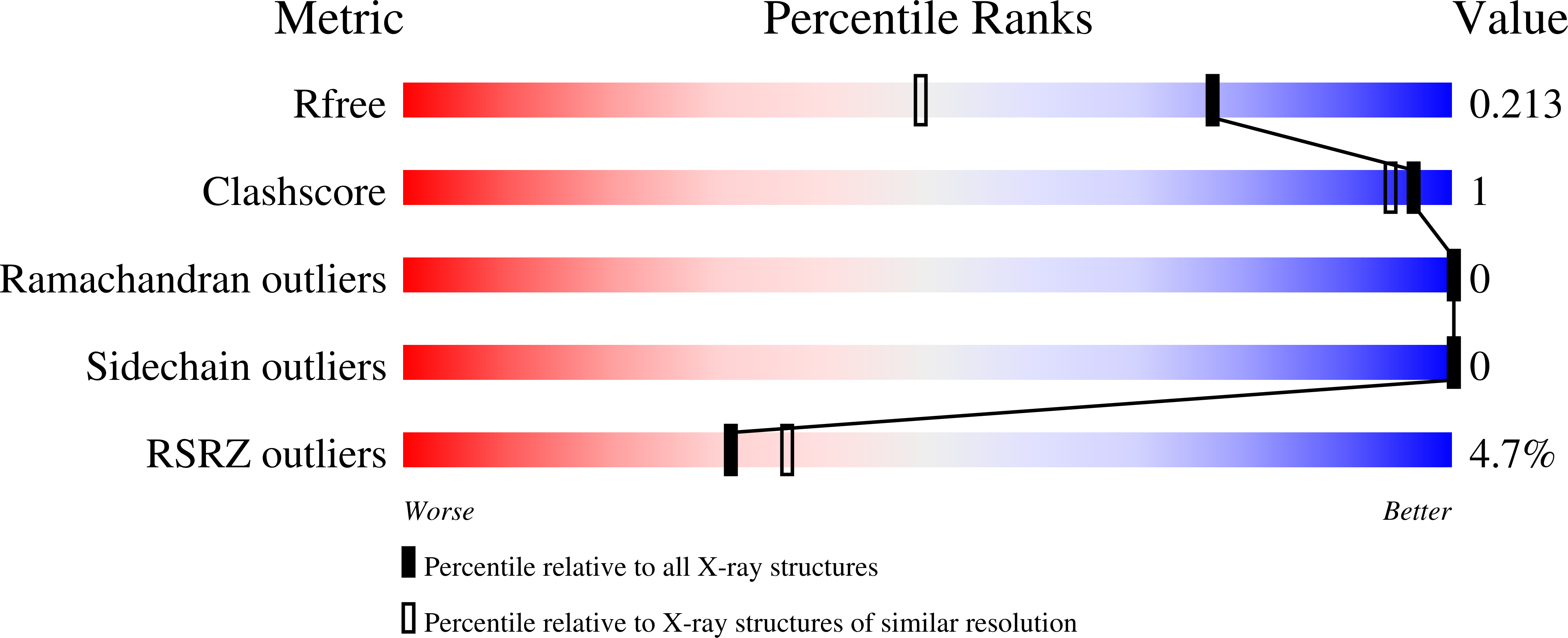
Elucidating the glycan-binding specificity and structure of Cucumis melo agglutinin, a new R-type lectin.
Lundstrom, J., Gillon, E., Chazalet, V., Kerekes, N., Di Maio, A., Feizi, T., Liu, Y., Varrot, A., Bojar, D.(2024) Beilstein J Org Chem 20: 306-320
- PubMed: 38410776
- DOI: https://doi.org/10.3762/bjoc.20.31
- Primary Citation of Related Structures:
8R8A, 8R8C - PubMed Abstract:
Plant lectins have garnered attention for their roles as laboratory probes and potential therapeutics. Here, we report the discovery and characterization of Cucumis melo agglutinin (CMA1), a new R-type lectin from melon. Our findings reveal CMA1's unique glycan-binding profile, mechanistically explained by its 3D structure, augmenting our understanding of R-type lectins. We expressed CMA1 recombinantly and assessed its binding specificity using multiple glycan arrays, covering 1,046 unique sequences. This resulted in a complex binding profile, strongly preferring C2-substituted, beta-linked galactose (both GalNAc and Fuca1-2Gal), which we contrasted with the established R-type lectin Ricinus communis agglutinin 1 (RCA1). We also report binding of specific glycosaminoglycan subtypes and a general enhancement of binding by sulfation. Further validation using agglutination, thermal shift assays, and surface plasmon resonance confirmed and quantified this binding specificity in solution. Finally, we solved the high-resolution structure of the CMA1 N-terminal domain using X-ray crystallography, supporting our functional findings at the molecular level. Our study provides a comprehensive understanding of CMA1, laying the groundwork for further exploration of its biological and therapeutic potential.
Organizational Affiliation:
Department of Chemistry and Molecular Biology, University of Gothenburg, Medicinaregatan 7B, 413 90 Gothenburg, Sweden.