Diflunisal versus tafamidis on neuropathy and cardiomyopathy in hereditary transthyretin amyloidosis.
Chao, C.C., Tzeng, S.R., Chiang, M.C., Hsueh, H.W., Hsieh, W.J., Chao, Y.C., Cheng, M.F., Lin, Y.H., Su, M.Y., Huang, C.H., Wang, Y.S., Hsieh, M.F., Tseng, P.H., Hsieh, S.T.(2024) Ann Clin Transl Neurol 11: 2426-2438
- PubMed: 39096004
- DOI: https://doi.org/10.1002/acn3.52158
- Primary Citation of Related Structures:
8YQD - PubMed Abstract:
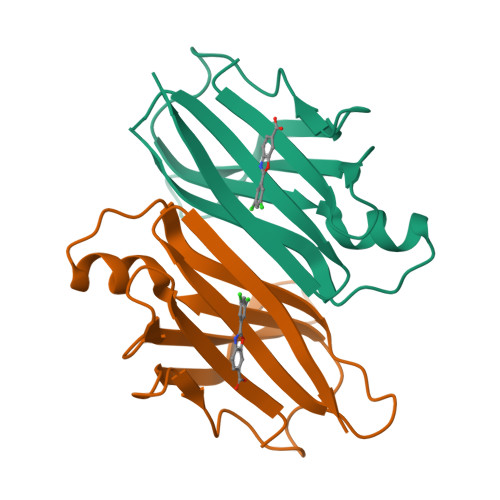
Hereditary transthyretin (TTR) amyloidosis (ATTRv) is frequently complicated by polyneuropathy (ATTRv-PN) and cardiomyopathy (ATTRv-CM). The long-term efficacy of diflunisal on both polyneuropathy and cardiomyopathy in ATTRv patients, especially those with non-V30M genotypes, has not been fully investigated and compared with that of tafamidis. We compared the structural and biochemical characteristics of A97S-TTR complexed with tafamidis with those of diflunisal, and prospectively followed up and compared the progression of polyneuropathy and cardiomyopathy between ATTRv-PN patients taking diflunisal and those taking tafamidis. Both diflunisal and tafamidis effectively bind to the two thyroxine-binding sites at the A97S-TTR dimer-dimer interface and equally and almost sufficiently reduce amyloid fibril formation. Thirty-five ATTRv-PN patients receiving diflunisal and 22 patients receiving tafamidis were enrolled. Compared with no treatment, diflunisal treatment significantly delayed the transition of FAP Stage 1 to 2 and Stage 2 to 3 and decreased the deterioration in parameters of the ulnar nerve conduction study (NCS). The progression of FAP stage or NCS parameters did not differ between patients treated with diflunisal and those treated with tafamidis. Both diflunisal and tafamidis treatments significantly decreased radiotracer uptake on 99m Tc-PYP SPECT and stabilized cardiac wall thickness and blood pro-B-type natriuretic peptide levels. No significant adverse events occurred during diflunisal or tafamidis treatment. The binding patterns of both tafamidis and diflunisal to A97S-TTR closely resembled those observed in the wild type. Diflunisal can effectively delay the progression of polyneuropathy and cardiomyopathy with similar efficacy to tafamidis and may become a cost-effective alternative treatment for late-onset ATTRv-PN.
Organizational Affiliation:
Department of Neurology, National Taiwan University Hospital, Taipei, Taiwan.