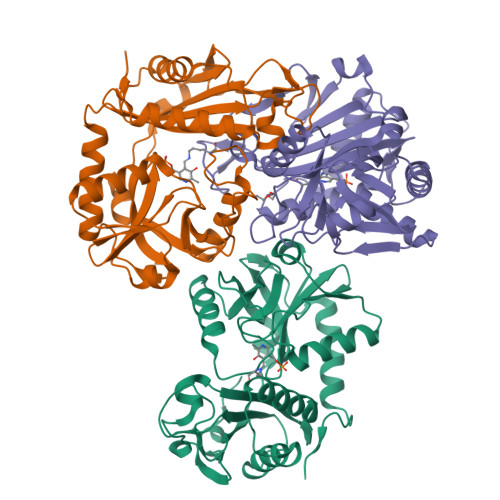
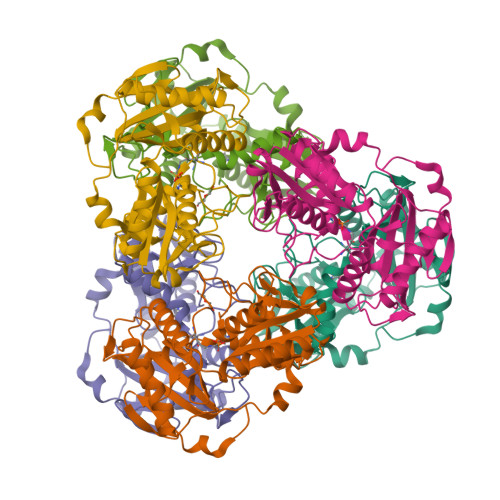
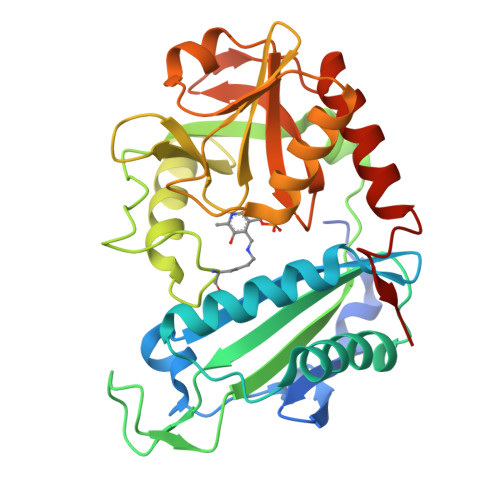
Protein of unknown function from variovorax paradoxus with amino acid substitution n174k is able to form schiff base with plp molecule
Ilyasov, I., Minyaev, M., Rakitina, T., Bakunova, A., Popov, V., Bezsudnova, E., Boyko, K.(2024) Кристаллография 69: 981-986