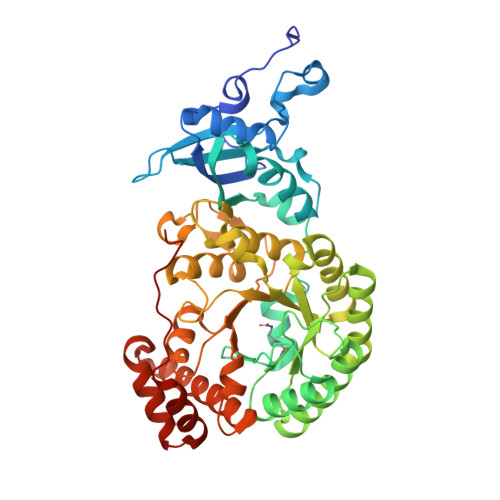
The structure of Mn(II)-bound Rubisco from Spinacia oleracea.
Voland, R.W., Coleman, R.E., Lancaster, K.M.(2024) J Inorg Biochem 260: 112682-112682
- PubMed: 39094246
- DOI: https://doi.org/10.1016/j.jinorgbio.2024.112682
- Primary Citation of Related Structures:
9CQ5 - PubMed Abstract:
The rate of photosynthesis and, thus, CO 2 fixation, is limited by the rate of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Not only does Rubisco have a relatively low catalytic rate, but it also is promiscuous regarding the metal identity in the active site of the large subunit. In Nature, Rubisco binds either Mg(II) or Mn(II), depending on the chloroplastic ratio of these metal ions; most studies performed with Rubisco have focused on Mg-bound Rubisco. Herein, we report the first crystal structure of a Mn-bound Rubisco, and we compare its structural properties to those of its Mg-bound analogues.
Organizational Affiliation:
Department of Chemistry and Chemical Biology Cornell University, Baker Laboratory, 162 Sciences Drive, Ithaca, NY 14853, USA.