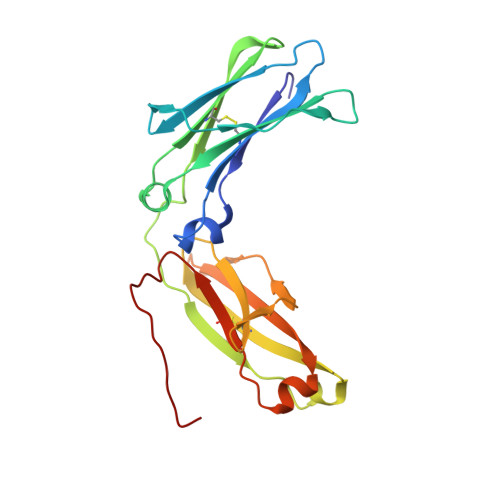
Generation and Characterization of an IgG4 Monomeric Fc Platform.
Shan, L., Colazet, M., Rosenthal, K.L., Yu, X.Q., Bee, J.S., Ferguson, A., Damschroder, M.M., Wu, H., Dall'Acqua, W.F., Tsui, P., Oganesyan, V.(2016) PLoS One 11: e0160345-e0160345
- PubMed: 27479095
- DOI: https://doi.org/10.1371/journal.pone.0160345
- Primary Citation of Related Structures:
5HVW - PubMed Abstract:
The immunoglobulin Fc region is a homodimer consisted of two sets of CH2 and CH3 domains and has been exploited to generate two-arm protein fusions with high expression yields, simplified purification processes and extended serum half-life. However, attempts to generate one-arm fusion proteins with monomeric Fc, with one set of CH2 and CH3 domains, are often plagued with challenges such as weakened binding to FcRn or partial monomer formation. Here, we demonstrate the generation of a stable IgG4 Fc monomer with a unique combination of mutations at the CH3-CH3 interface using rational design combined with in vitro evolution methodologies. In addition to size-exclusion chromatography and analytical ultracentrifugation, we used multi-angle light scattering (MALS) to show that the engineered Fc monomer exhibits excellent monodispersity. Furthermore, crystal structure analysis (PDB ID: 5HVW) reveals monomeric properties supported by disrupted interactions at the CH3-CH3 interface. Monomeric Fc fusions with Fab or scFv achieved FcRn binding and serum half-life comparable to wildtype IgG. These results demonstrate that this monomeric IgG4 Fc is a promising therapeutic platform to extend the serum half-life of proteins in a monovalent format.
Organizational Affiliation:
Antibody Discovery and Protein Engineering, MedImmune, Gaithersburg, Maryland, United States of America.