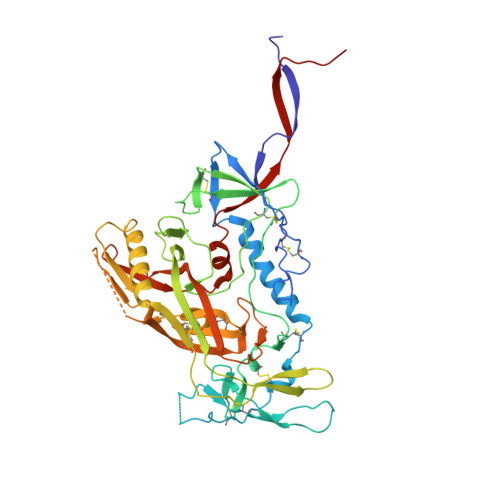
Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529.
Pancera, M., Lai, Y.T., Bylund, T., Druz, A., Narpala, S., O'Dell, S., Schon, A., Bailer, R.T., Chuang, G.Y., Geng, H., Louder, M.K., Rawi, R., Soumana, D.I., Finzi, A., Herschhorn, A., Madani, N., Sodroski, J., Freire, E., Langley, D.R., Mascola, J.R., McDermott, A.B., Kwong, P.D.(2017) Nat Chem Biol 13: 1115-1122
- PubMed: 28825711
- DOI: https://doi.org/10.1038/nchembio.2460
- Primary Citation of Related Structures:
5U7M, 5U7O - PubMed Abstract:
The HIV-1 envelope (Env) spike is a conformational machine that transitions between prefusion (closed, CD4- and CCR5-bound) and postfusion states to facilitate HIV-1 entry into cells. Although the prefusion closed conformation is a potential target for inhibition, development of small-molecule leads has been stymied by difficulties in obtaining structural information. Here, we report crystal structures at 3.8-Å resolution of an HIV-1-Env trimer with BMS-378806 and a derivative BMS-626529 for which a prodrug version is currently in Phase III clinical trials. Both lead candidates recognized an induced binding pocket that was mostly excluded from solvent and comprised of Env elements from a conserved helix and the β20-21 hairpin. In both structures, the β20-21 region assumed a conformation distinct from prefusion-closed and CD4-bound states. Together with biophysical and antigenicity characterizations, the structures illuminate the allosteric and competitive mechanisms by which these small-molecule leads inhibit CD4-induced structural changes in Env.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.