The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Mathews, I.I., Allison, K., Robbins, T., Lyubimov, A.Y., Uervirojnangkoorn, M., Brunger, A.T., Khosla, C., DeMirci, H., McPhillips, S.E., Hollenbeck, M., Soltis, M., Cohen, A.E.(2017) Biochemistry 56: 4751-4756
- PubMed: 28832129
- DOI: https://doi.org/10.1021/acs.biochem.7b00711
- Primary Citation of Related Structures:
6APF, 6APG, 6APK, 6APM - PubMed Abstract:
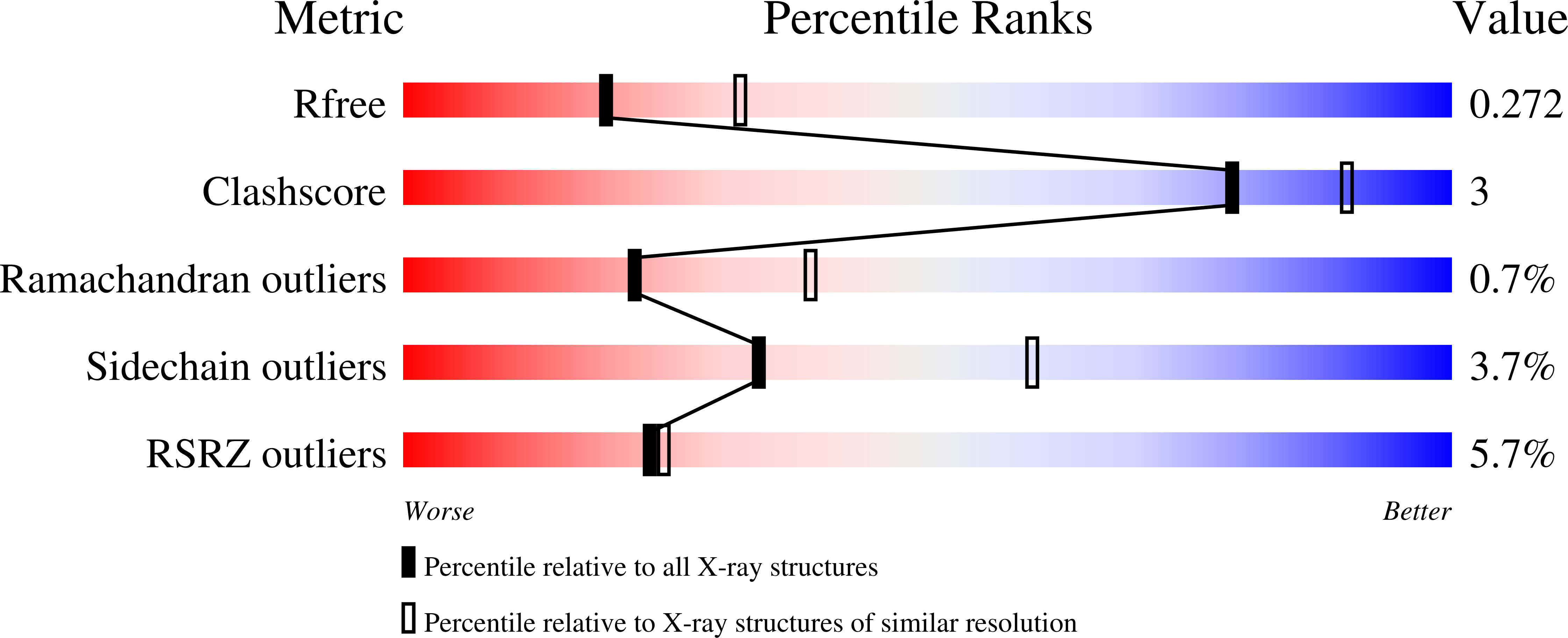
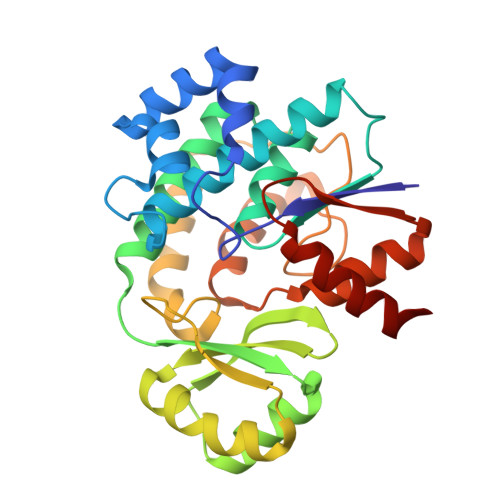
The crystal structure of the trans-acyltransferase (AT) from the disorazole polyketide synthase (PKS) was determined at room temperature to a resolution of 2.5 Å using a new method for the direct delivery of the sample into an X-ray free-electron laser. A novel sample extractor efficiently delivered limited quantities of microcrystals directly from the native crystallization solution into the X-ray beam at room temperature. The AT structure revealed important catalytic features of this core PKS enzyme, including the occurrence of conformational changes around the active site. The implications of these conformational changes for polyketide synthase reaction dynamics are discussed.
Organizational Affiliation:
Stanford Synchrotron Radiation Lightsource , 2575 Sand Hill Road, Menlo Park, California 94025, United States.