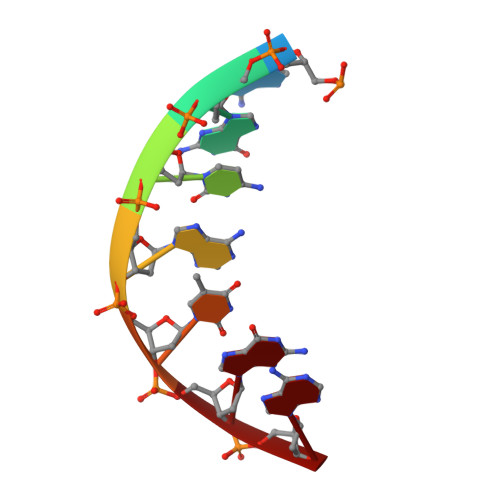
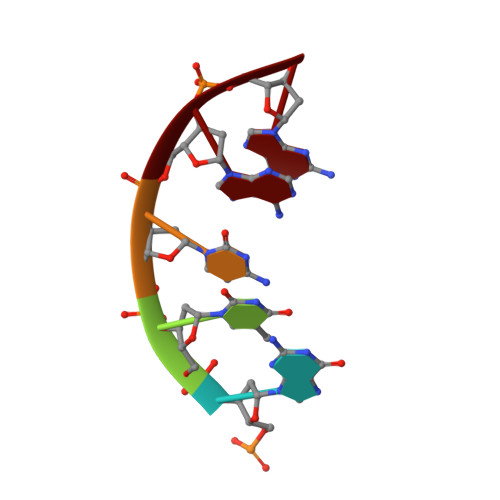
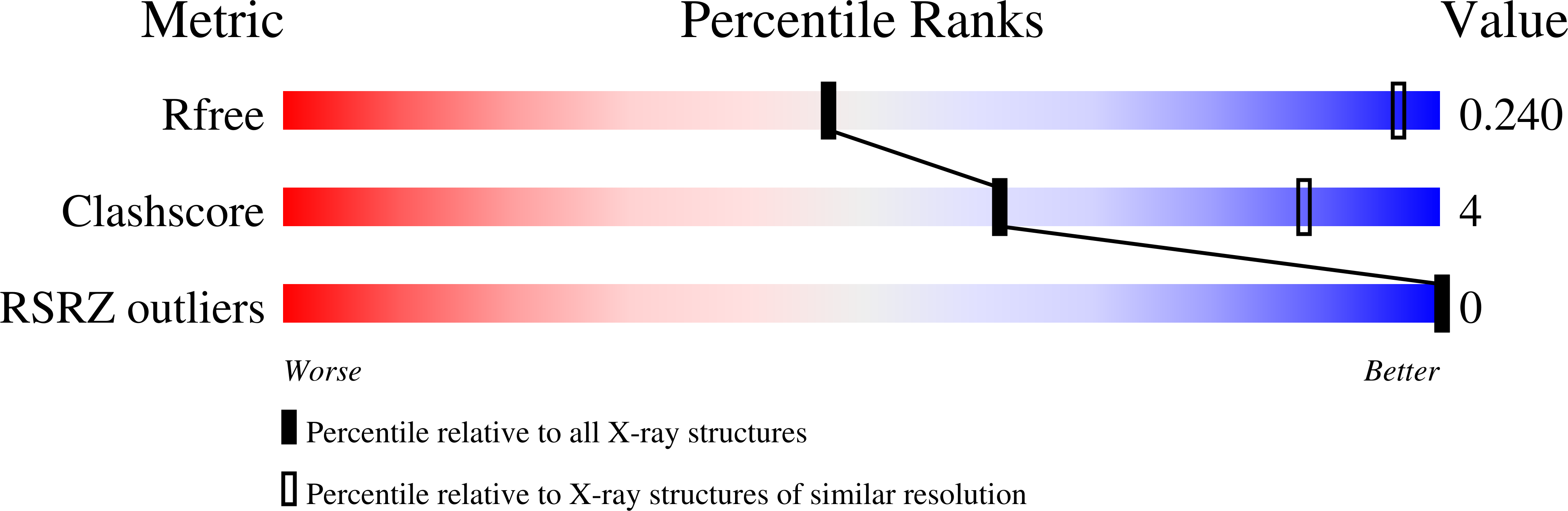A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Simmons, C.R., MacCulloch, T., Zhang, F., Liu, Y., Stephanopoulos, N., Yan, H.(2020) Angew Chem Int Ed Engl 59: 18619-18626
- PubMed: 32533629
- DOI: https://doi.org/10.1002/anie.202005505
- Primary Citation of Related Structures:
6U40, 6UDN, 6UEF, 6V6R, 6WJK - PubMed Abstract:
DNA is an ideal molecule for the construction of 3D crystals with tunable properties owing to its high programmability based on canonical Watson-Crick base pairing, with crystal assembly in all three dimensions facilitated by immobile Holliday junctions and sticky end cohesion. Despite the promise of these systems, only a handful of unique crystal scaffolds have been reported. Herein, we describe a new crystal system with a repeating sequence that mediates the assembly of a 3D scaffold via a series of Holliday junctions linked together with complementary sticky ends. By using an optimized junction sequence, we could determine a high-resolution (2.7 Å) structure containing R3 crystal symmetry, with a slight subsequent improvement (2.6 Å) using a modified sticky-end sequence. The immobile Holliday junction sequence allowed us to produce crystals that provided unprecedented atomic detail. In addition, we expanded the crystal cavities by 50 % by adding an additional helical turn between junctions, and we solved the structure to 4.5 Å resolution by molecular replacement.
Organizational Affiliation:
Biodesign Center for Molecular Design and Biomimetics, Arizona State University, USA.