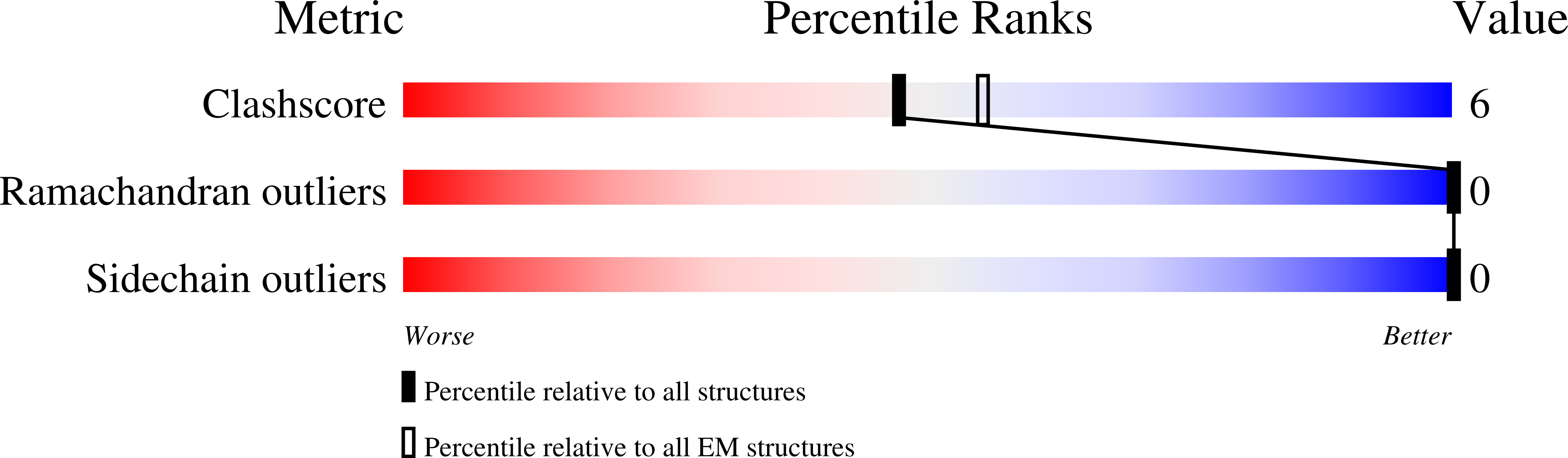The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
McKenna, M.J., Sim, S.I., Ordureau, A., Wei, L., Harper, J.W., Shao, S., Park, E.(2020) Science 369
- PubMed: 32973005
- DOI: https://doi.org/10.1126/science.abc5809
- Primary Citation of Related Structures:
6XMP, 6XMQ, 6XMS, 6XMT, 6XMU - PubMed Abstract:
Organelle identity depends on protein composition. How mistargeted proteins are selectively recognized and removed from organelles is incompletely understood. Here, we found that the orphan P5A-adenosine triphosphatase (ATPase) transporter ATP13A1 (Spf1 in yeast) directly interacted with the transmembrane segment (TM) of mitochondrial tail-anchored proteins. P5A-ATPase activity mediated the extraction of mistargeted proteins from the endoplasmic reticulum (ER). Cryo-electron microscopy structures of Saccharomyces cerevisiae Spf1 revealed a large, membrane-accessible substrate-binding pocket that alternately faced the ER lumen and cytosol and an endogenous substrate resembling an α-helical TM. Our results indicate that the P5A-ATPase could dislocate misinserted hydrophobic helices flanked by short basic segments from the ER. TM dislocation by the P5A-ATPase establishes an additional class of P-type ATPase substrates and may correct mistakes in protein targeting or topogenesis.
Organizational Affiliation:
Department of Cell Biology, Blavatnik Institute, Harvard Medical School, Boston, MA 02115, USA.