A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor gamma Binding Properties and Biological Activities.
Peiretti, F., Montanari, R., Capelli, D., Bonardo, B., Colson, C., Amri, E.Z., Grimaldi, M., Balaguer, P., Ito, K., Roeder, R.G., Pochetti, G., Brunel, J.M.(2020) J Med Chem 63: 13124-13139
- PubMed: 33142057
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01555
- Primary Citation of Related Structures:
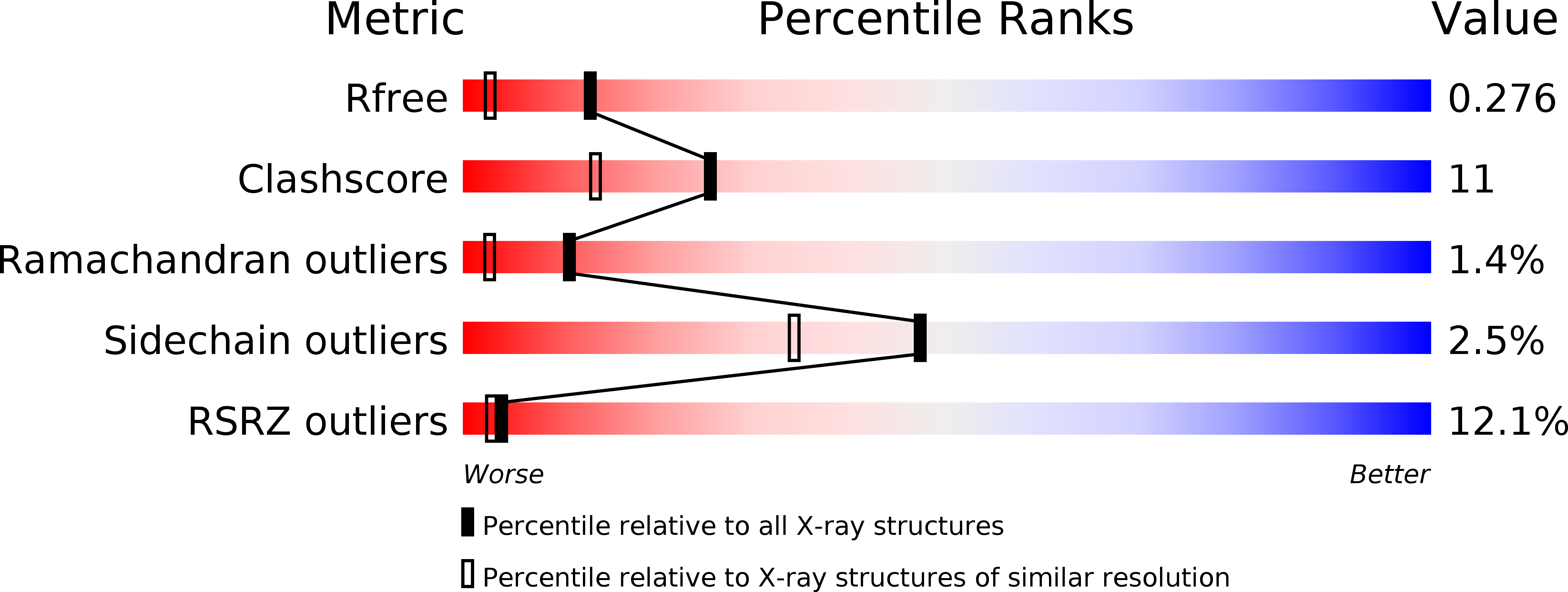
6QJ5, 6ZLY - PubMed Abstract:
A proprietary library of novel N -aryl-substituted amino acid derivatives bearing a hydroxamate head group allowed the identification of compound 3a that possesses weak proadipogenic and peroxisome proliferator-activated receptor γ (PPARγ) activating properties. The systematic optimization of 3a , in order to improve its PPARγ agonist activity, led to the synthesis of compound 7j ( N -aryl-substituted valine derivative) that possesses dual PPARγ/PPARα agonistic activity. Structural and kinetic analyses reveal that 7j occupies the typical ligand binding domain of the PPARγ agonists with, however, a unique high-affinity binding mode. Furthermore, 7j is highly effective in preventing cyclin-dependent kinase 5-mediated phosphorylation of PPARγ serine 273. Although less proadipogenic than rosiglitazone, 7j significantly increases adipocyte insulin-stimulated glucose uptake and efficiently promotes white-to-brown adipocyte conversion. In addition, 7j prevents oleic acid-induced lipid accumulation in hepatoma cells. The unique biochemical properties and biological activities of compound 7j suggest that it would be a promising candidate for the development of compounds to reduce insulin resistance, obesity, and nonalcoholic fatty liver disease.
Organizational Affiliation:
Aix Marseille University, INSERM, INRAE, C2VN, 13385 Marseille, France.