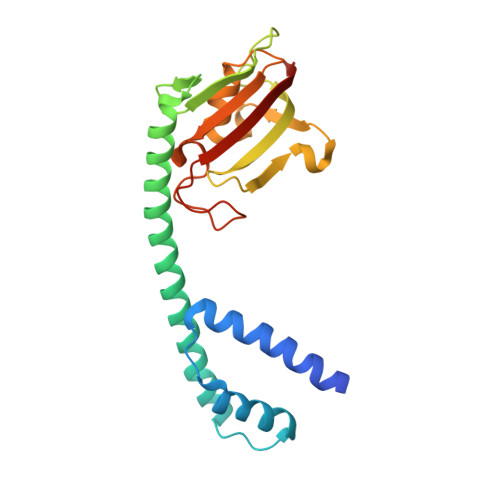
Crystal structure of Mip, a prolylisomerase from Legionella pneumophila
Riboldi-Tunnicliffe, A., Konig, B., Jessen, S., Weiss, M.S., Rahfeld, J., Hacker, J., Fischer, G., Hilgenfeld, R.(2001) Nat Struct Biol 8: 779-783
- PubMed: 11524681
- DOI: https://doi.org/10.1038/nsb0901-779
- Primary Citation of Related Structures:
1FD9 - PubMed Abstract:
The human pathogen Legionella pneumophila, the etiological agent of the severe and often fatal Legionnaires' disease, produces a major virulence factor, termed 'macrophage infectivity potentiator protein' (Mip), that is necessary for optimal multiplication of the bacteria within human alveolar macrophages. Mip exhibits a peptidyl prolyl cis-trans isomerase (PPIase) activity, which appears to be important for infection. Here we report the 2.4 A crystal structure of the Mip protein from L. pneumophila Philadelphia 1 and the 3.2 A crystal structure of its complex with the drug FK506. Each monomer of the homodimeric protein consists of an N-terminal dimerization module, a long (65 A) connecting alpha-helix and a C-terminal PPIase domain exhibiting similarity to human FK506-binding protein. In view of the recent significant increase in the number of reported cases of Legionnaires' disease and other intracellular infections, these structural results are of prime interest for the design of new drugs directed against Mip proteins of intracellular pathogens.
- Institute of Molecular Biotechnology, Beutenbergstr. 11, D-07745 Jena, Germany.
Organizational Affiliation: