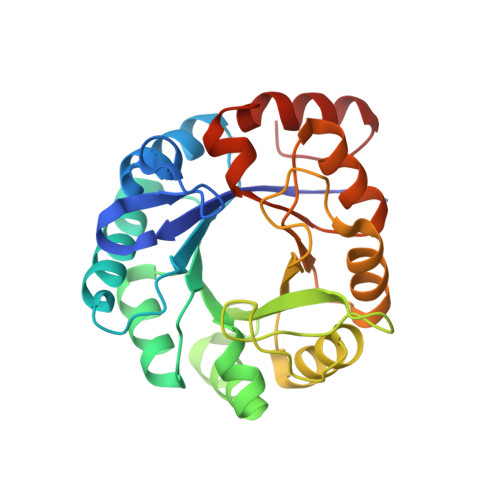
Structure of Hisf, a Histidine Biosynthetic Protein from Pyrobaculum Aerophilum
Banfield, M.J., Lott, J.S., Arcus, V., Mccarthy, A.A., Baker, E.N.(2001) Acta Crystallogr D Biol Crystallogr 57: 1518
- PubMed: 11679715
- DOI: https://doi.org/10.1107/s0907444901012604
- Primary Citation of Related Structures:
1H5Y - PubMed Abstract:
HisF (imidazole glycerol phosphate synthase) is an important branch-point enzyme in the histidine biosynthetic pathway of microorganisms. Because of its potential relevance for structure-based drug design, the crystal structure of HisF from the hyperthermophilic archaeon Pyrobaculum aerophilum has been determined. The structure was determined by molecular replacement and refined at 2.0 A resolution to a crystallographic R factor of 20.6% and a free R of 22.7%. The structure adopts a classic (beta/alpha)(8) barrel fold and has networks of surface salt bridges that may contribute to thermostability. The active site is marked out by the presence of two bound phosphate ions and two glycerol molecules that delineate a long groove at one end of the (beta/alpha)(8) barrel. The two phosphate ions, 17 A apart, are bound to sequence-conserved structural motifs that seem likely to provide much of the specificity for the two phosphate groups of the HisF substrate. The two glycerol molecules bind in the vicinity of other sequence-conserved residues that are likely to be involved in binding and/or catalysis. Comparisons with the homologous HisF from Thermatoga maritima reveal a displaced loop that may serve as a lid over the active site.
- School of Biological Sciences, University of Auckland, Private Bag 92019, Auckland, New Zealand.
Organizational Affiliation: