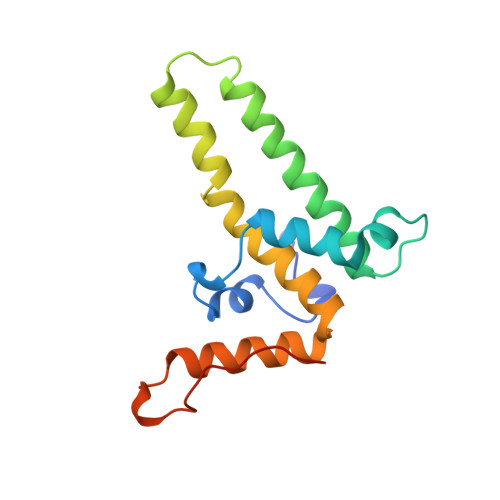
The crystal structure of the human hepatitis B virus capsid.
Wynne, S.A., Crowther, R.A., Leslie, A.G.(1999) Mol Cell 3: 771-780
- PubMed: 10394365
- DOI: https://doi.org/10.1016/s1097-2765(01)80009-5
- Primary Citation of Related Structures:
1QGT - PubMed Abstract:
Hepatitis B is a small enveloped DNA virus that poses a major hazard to human health. The crystal structure of the T = 4 capsid has been solved at 3.3 A resolution, revealing a largely helical protein fold that is unusual for icosahedral viruses. The monomer fold is stabilized by a hydrophobic core that is highly conserved among human viral variants. Association of two amphipathic alpha-helical hairpins results in formation of a dimer with a four-helix bundle as the major central feature. The capsid is assembled from dimers via interactions involving a highly conserved region near the C terminus of the truncated protein used for crystallization. The major immunodominant region lies at the tips of the alpha-helical hairpins that form spikes on the capsid surface.
- Medical Research Council, Laboratory of Molecular Biology, Cambridge, United Kingdom.
Organizational Affiliation: