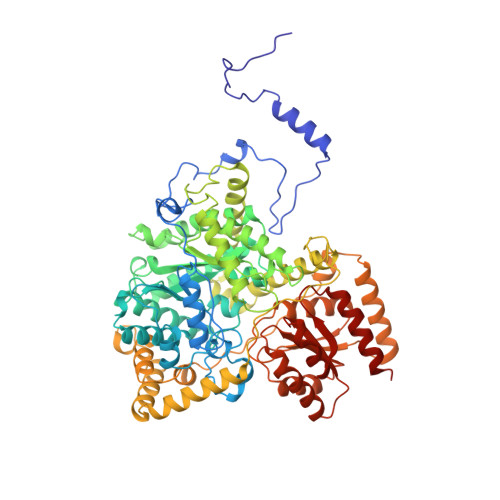
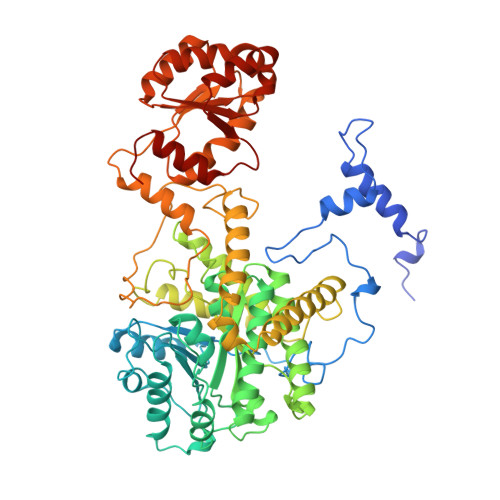
How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution.
Mancia, F., Keep, N.H., Nakagawa, A., Leadlay, P.F., McSweeney, S., Rasmussen, B., Bosecke, P., Diat, O., Evans, P.R.(1996) Structure 4: 339-350
- PubMed: 8805541
- DOI: https://doi.org/10.1016/s0969-2126(96)00037-8
- Primary Citation of Related Structures:
1REQ - PubMed Abstract:
The enzyme methylmalonyl-coenzyme A (CoA) mutase, an alphabeta heterodimer of 150 kDa, is a member of a class of enzymes that uses coenzyme B12 (adenosylcobalamin) as a cofactor. The enzyme induces the formation of an adenosyl radical from the cofactor. This radical then initiates a free-radical rearrangement of its substrate, succinyl-CoA, to methylmalonyl-CoA. Reported here is the crystal structure at 2 A resolution of methylmalonyl-CoA mutase from Propionibacterium shermanii in complex with coenzyme B12 and with the partial substrate desulpho-CoA (lacking the succinyl group and the sulphur atom of the substrate). The coenzyme is bound by a domain which shares a similar fold to those of flavodoxin and the B12-binding domain of methylcobalamin-dependent methionine synthase. The cobalt atom is coordinated, via a long bond, to a histidine from the protein. The partial substrate is bound along the axis of a (beta/alpha)8 TIM barrel domain. The histidine-cobalt distance is very long (2.5 A compared with 1.95-2.2 A in free cobalamins), suggesting that the enzyme positions the histidine in order to weaken the metal-carbon bond of the cofactor and favour the formation of the initial radical species. The active site is deeply buried, and the only access to it is through a narrow tunnel along the axis of the TIM barrel domain.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK.
Organizational Affiliation: