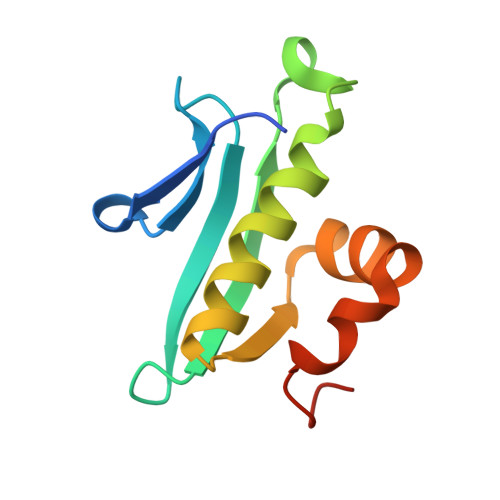
Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Tyler, R.C., Bitto, E., Berndsen, C.E., Bingman, C.A., Singh, S., Lee, M.S., Wesenberg, G.E., Denu, J.M., Phillips Jr., G.N., Markley, J.L.(2006) Biochemistry 45: 14325-14336
- PubMed: 17128971
- DOI: https://doi.org/10.1021/bi0612059
- Primary Citation of Related Structures:
1XMT, 2EVN, 2IL4 - PubMed Abstract:
We describe X-ray crystal and NMR solution structures of the protein coded for by Arabidopsis thaliana gene At1g77540.1 (At1g77540). The crystal structure was determined to 1.15 A with an R factor of 14.9% (Rfree = 17.0%) by multiple-wavelength anomalous diffraction using sodium bromide derivatized crystals. The ensemble of NMR conformers was determined with protein samples labeled with 15N and 13C + 15N. The X-ray structure and NMR ensemble were closely similar with rmsd 1.4 A for residues 8-93. At1g77540 was found to adopt a fold similar to that of GCN5-related N-acetyltransferases. Enzymatic activity assays established that At1g77540 possesses weak acetyltransferase activity against histones H3 and H4. Chemical shift perturbations observed in 15N-HSQC spectra upon the addition of CoA indicated that the cofactor binds and identified its binding site. The molecular details of this interaction were further elucidated by solving the X-ray structure of the At1g77540-CoA complex. This work establishes that the domain family COG2388 represents a novel class of acetyltransferase and provides insight into possible mechanistic roles of the conserved Cys76 and His41 residues of this family.
- Center for Eukaryotic Structural Genomics, Biochemistry Department, College of Agricultural and Life Sciences, University of Wisconsin-Madison, 433 Babcock Drive, Madison, Wisconsin 53706-1544, USA.
Organizational Affiliation: