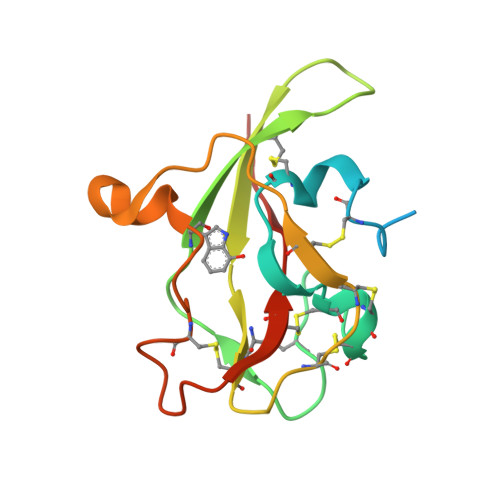
Atomic description of an enzyme reaction dominated by proton tunneling
Masgrau, L., Roujeinikova, A., Johannissen, L.O., Hothi, P., Basran, J., Ranaghan, K.E., Mulholland, A.J., Sutcliffe, M.J., Scrutton, N.S., Leys, D.(2006) Science 312: 237-241
- PubMed: 16614214
- DOI: https://doi.org/10.1126/science.1126002
- Primary Citation of Related Structures:
2AGL, 2AGW, 2AGX, 2AGY, 2AGZ, 2AH0, 2AH1 - PubMed Abstract:
We present an atomic-level description of the reaction chemistry of an enzyme-catalyzed reaction dominated by proton tunneling. By solving structures of reaction intermediates at near-atomic resolution, we have identified the reaction pathway for tryptamine oxidation by aromatic amine dehydrogenase. Combining experiment and computer simulation, we show proton transfer occurs predominantly to oxygen O2 of Asp(128)beta in a reaction dominated by tunneling over approximately 0.6 angstroms. The role of long-range coupled motions in promoting tunneling is controversial. We show that, in this enzyme system, tunneling is promoted by a short-range motion modulating proton-acceptor distance and no long-range coupled motion is required.
Organizational Affiliation:
Manchester Interdisciplinary Biocentre, University of Manchester, Jackson's Mill, Post Office Box 88, Manchester M60 1QD, UK.