Novel features of the rotary catalytic mechanism revealed in the structure of yeast F(1) ATPase.
Kabaleeswaran, V., Puri, N., Walker, J.E., Leslie, A.G., Mueller, D.M.(2006) EMBO J 25: 5433-5442
- PubMed: 17082766
- DOI: https://doi.org/10.1038/sj.emboj.7601410
- Primary Citation of Related Structures:
2HLD - PubMed Abstract:
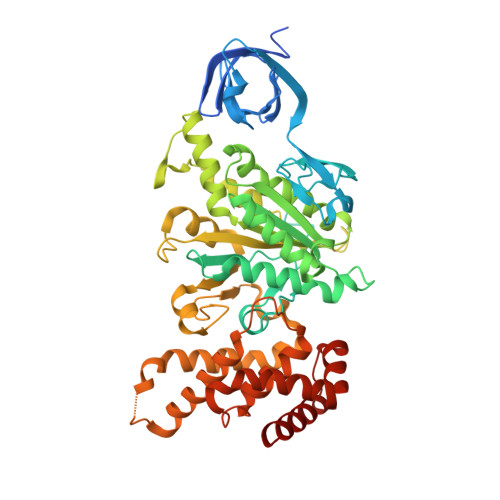
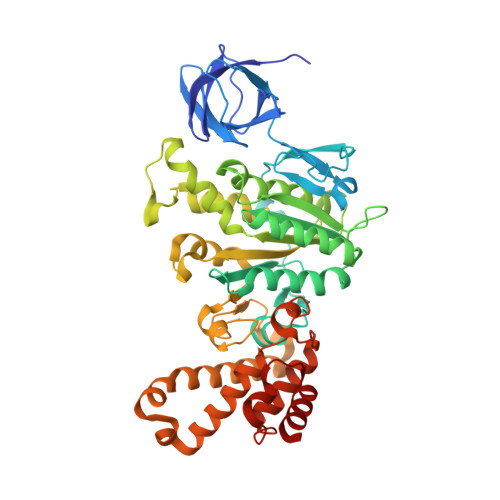
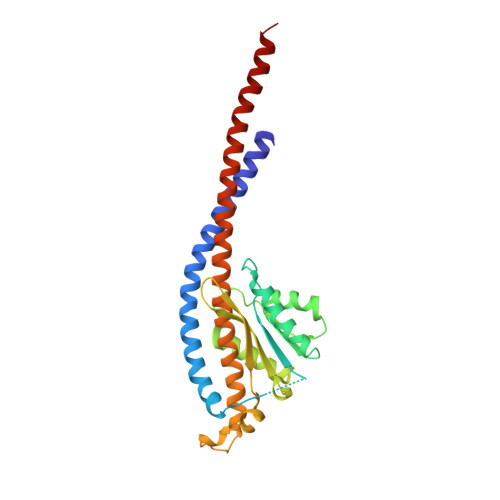
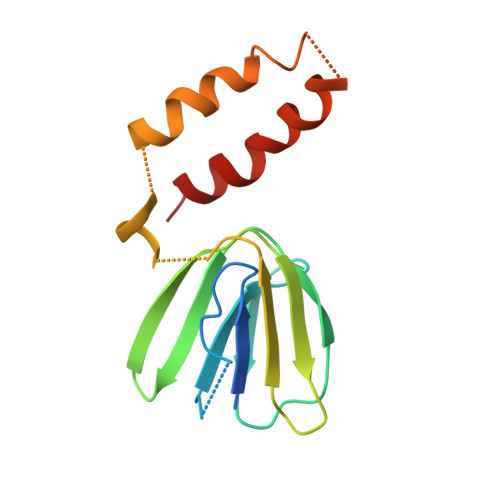
The crystal structure of yeast mitochondrial F(1) ATPase contains three independent copies of the complex, two of which have similar conformations while the third differs in the position of the central stalk relative to the alpha(3)beta(3) sub-assembly. All three copies display very similar asymmetric features to those observed for the bovine enzyme, but the yeast F(1) ATPase structures provide novel information. In particular, the active site that binds ADP in bovine F(1) ATPase has an ATP analog bound and therefore this structure does not represent the ADP-inhibited form. In addition, one of the complexes binds phosphate in the nucleotide-free catalytic site, and comparison with other structures provides a picture of the movement of the phosphate group during initial binding and subsequent catalysis. The shifts in position of the central stalk between two of the three copies of yeast F(1) ATPase and when these structures are compared to those of the bovine enzyme give new insight into the conformational changes that take place during rotational catalysis.
- Department of Biochemistry & Molecular Biology, The Chicago Medical School, Rosalind Franklin University of Medicine and Science, North Chicago, IL 60064, USA.
Organizational Affiliation: