Structure and Function of the Escherichia coli Protein YmgB: A Protein Critical for Biofilm Formation and Acid-resistance.
Lee, J., Page, R., Garcia-Contreras, R., Palermino, J.M., Zhang, X.S., Doshi, O., Wood, T.K., Peti, W.(2007) J Mol Biol 373: 11-26
- PubMed: 17765265
- DOI: https://doi.org/10.1016/j.jmb.2007.07.037
- Primary Citation of Related Structures:
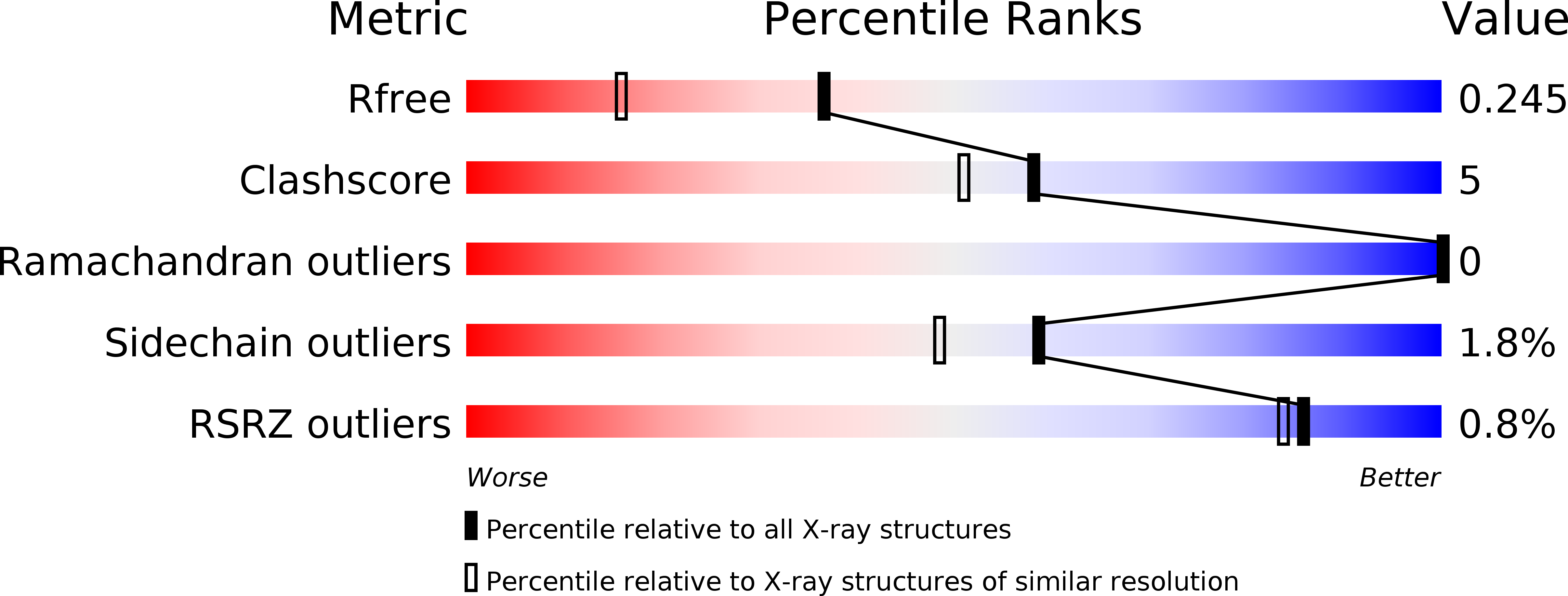
2OXL - PubMed Abstract:
The Escherichia coli gene cluster ymgABC was identified in transcriptome studies to have a role in biofilm development and stability. In this study, we showed that YmgB represses biofilm formation in rich medium containing glucose, decreases cellular motility, and protects the cell from acid indicating that YmgB has a major role in acid-resistance in E. coli. Our data show that these phenotypes are potentially mediated through interactions with the important cell signal indole. In addition, gel mobility-shift assays suggest that YmgB may be a non-specific DNA-binding protein. Using nickel-enrichment DNA microarrays, we showed that YmgB binds, either directly or indirectly, via a probable ligand, genes important for biofilm formation. To advance our understanding of the function of YmgB, we used X-ray crystallography to solve the structure of the protein to 1.8 A resolution. YmgB is a biological dimer that is structurally homologous to the E. coli gene regulatory protein Hha, despite having only 5% sequence identity. This supports our DNA microarray data showing that YmgB is a gene regulatory protein. Therefore, this protein, which clearly has a critical role in acid-resistance in E. coli, has been renamed as AriR for regulator of acid resistance influenced by indole.
Organizational Affiliation:
Artie McFerrin Department of Chemical Engineering, Texas A & M University, College Station, TX 77843-3122, USA.