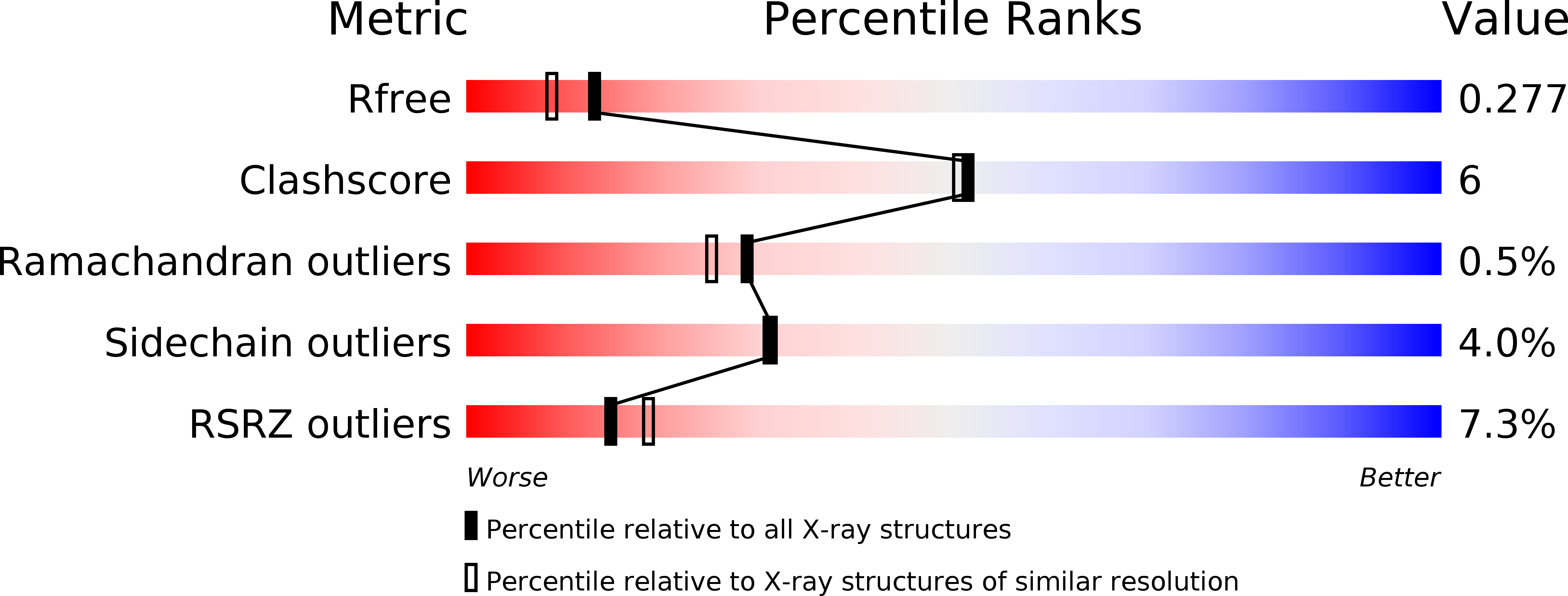
Human Oga Binds Substrates in a Conserved Peptide Recognition Groove.
Schimpl, M., Schuttelkopf, A.W., Borodkin, V.S., Van Aalten, D.M.F.(2010) Biochem J 432: 1
- PubMed: 20863279
- DOI: https://doi.org/10.1042/BJ20101338
- Primary Citation of Related Structures:
2XSA, 2XSB - PubMed Abstract:
Modification of cellular proteins with O-GlcNAc (O-linked N-acetylglucosamine) competes with protein phosphorylation and regulates a plethora of cellular processes. O-GlcNAcylation is orchestrated by two opposing enzymes, O-GlcNAc transferase and OGA (O-GlcNAcase or β-N-acetylglucosaminidase), which recognize their target proteins via as yet unidentified mechanisms. In the present study, we uncovered the first insights into the mechanism of substrate recognition by human OGA. The structure of a novel bacterial OGA orthologue reveals a putative substrate-binding groove, conserved in metazoan OGAs. Guided by this structure, conserved amino acids lining this groove in human OGA were mutated and the activity on three different substrate proteins [TAB1 (transforming growth factor-β-activated protein kinase 1-binding protein 1), FoxO1 (forkhead box O1) and CREB (cAMP-response-element-binding protein)] was tested in an in vitro deglycosylation assay. The results provide the first evidence that human OGA may possess a substrate-recognition mechanism that involves interactions with O-GlcNAcylated proteins beyond the GlcNAc-binding site, with possible implications for differential regulation of cycling of O-GlcNAc on different proteins.
Organizational Affiliation:
University of Dundee, Scotland, UK.