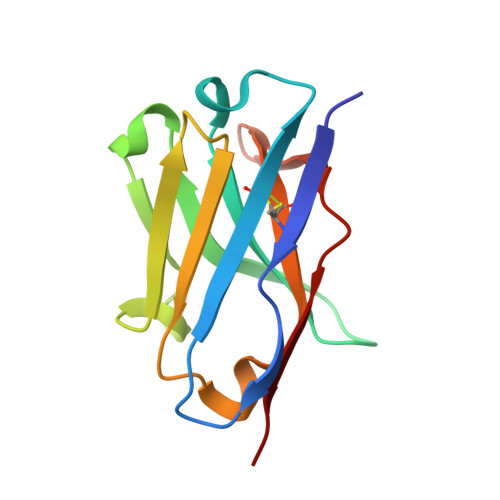
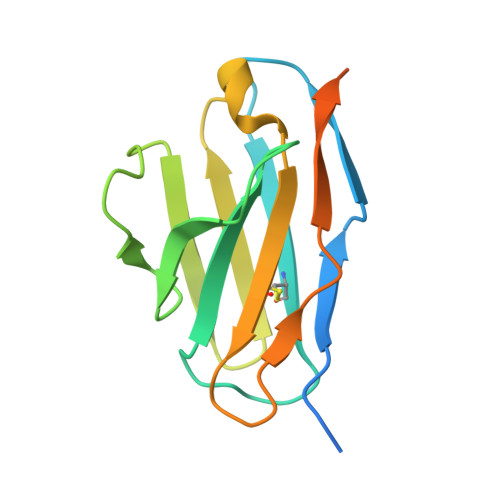
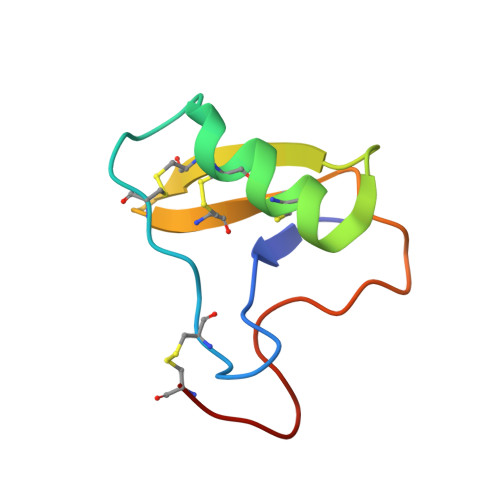
Structural Basis of Neutralization of the Major Toxic Component from the Scorpion Centruroides Noxius Hoffmann by a Human-Derived Single Chain Antibody Fragment.
Canul-Tec, J.C., Riano-Umbarila, L., Rudino-Pinera, E., Becerril, B., Possani, L.D., Torres-Larios, A.(2011) J Biol Chem 286: 20892
- PubMed: 21489992
- DOI: https://doi.org/10.1074/jbc.M111.238410
- Primary Citation of Related Structures:
2YBR, 2YC1 - PubMed Abstract:
It has previously been reported that several single-chain antibody fragments of human origin (scFv) neutralize the effects of two different scorpion venoms through interactions with the primary toxins of Centruroides noxius Hoffmann (Cn2) and Centruroides suffusus suffusus (Css2). Here we present the crystal structure of the complex formed between one scFv (9004G) and the Cn2 toxin, determined in two crystal forms at 2.5 and 1.9 Å resolution. A 15-residue span of the toxin is recognized by the antibody through a cleft formed by residues from five of the complementarity-determining regions of the scFv. Analysis of the interface of the complex reveals three features. First, the epitope of toxin Cn2 overlaps with essential residues for the binding of β-toxins to its Na(+) channel receptor site. Second, the putative recognition of Css2 involves mainly residues that are present in both Cn2 and Css2 toxins. Finally, the effect on the increase of affinity of previously reported key residues during the maturation process of different scFvs can be inferred from the structure. Taken together, these results provide the structural basis that explain the mechanism of the 9004G neutralizing activity and give insight into the process of directed evolution that gave rise to this family of neutralizing scFvs.
Organizational Affiliation:
Departamento de Medicina Molecular y Bioprocesos, Instituto de Biotecnología, Universidad Nacional Autónoma de México, Apartado Postal 510-3, Cuernavaca, Morelos 62210, USA.