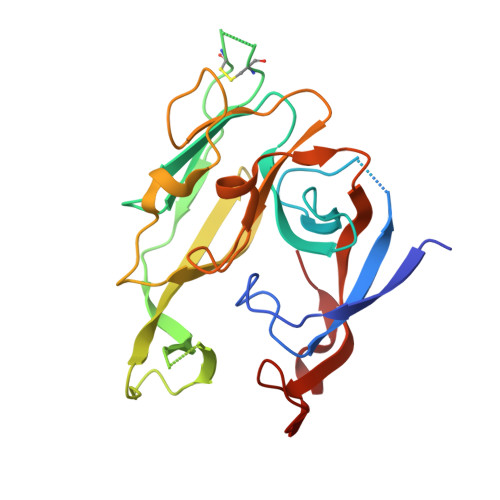
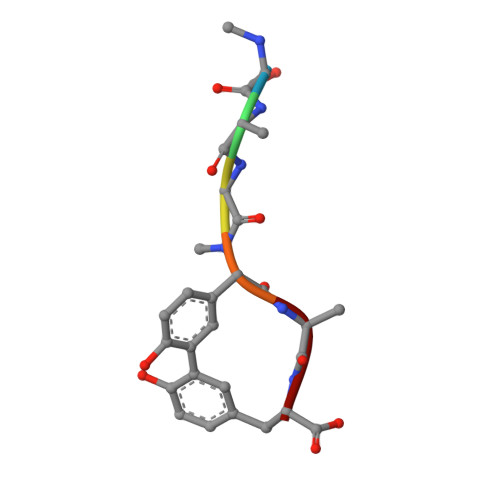
Crystallographic Analysis of Bacterial Signal Peptidase in Ternary Complex with Arylomycin A2 and a Beta-Sultam Inhibitor.
Luo, C., Roussel, P., Dreier, J., Page, M.G., Paetzel, M.(2009) Biochemistry 48: 8976
- PubMed: 19655811
- DOI: https://doi.org/10.1021/bi9009538
- Primary Citation of Related Structures:
3IIQ - PubMed Abstract:
Bacterial type I signal peptidase (SPase I), an essential membrane-bound endopeptidase with a unique Ser/Lys dyad mechanism, is being investigated as a potential novel antibiotic target. We present here binding and inhibition assays along with crystallographic data that shows that the lipohexapeptide-based natural product arylomycin A2 and the morpholino-beta-sultam derivative (BAL0019193) inhibit SPase I by binding to non-overlapping subsites near the catalytic center. The 2.0 A resolution crystal structure of the soluble catalytic domain of Escherichia coli SPase I (SPase I Delta2-75) in ternary complex with arylomycin A2 and BAL0019193 reveals the position of BAL0019193 adjacent to arylomycin A2 within the SPase I binding site. BAL0019193 binds in a noncovalent manner in close proximity to SPase I residues Ser88, Ser90, Lys145, Asn277, Ala279, and Glu307, as well as atom O45 of arylomycin A2. The binding mode of arylomycin A2 in this 2.0 A resolution ternary complex is compared to that seen in the previous 2.5 A resolution arylomycin A2-SPase cocrystal structure. This work contributes to our understanding of SPase I inhibitor/substrate recognition and should prove helpful in the further development of novel antibiotics based on the inhibition of SPase I.
- Department of Molecular Biology and Biochemistry, Simon Fraser University, South Science Building 8888 University Drive, Burnaby, British Columbia, V5A 1S6 Canada.
Organizational Affiliation: