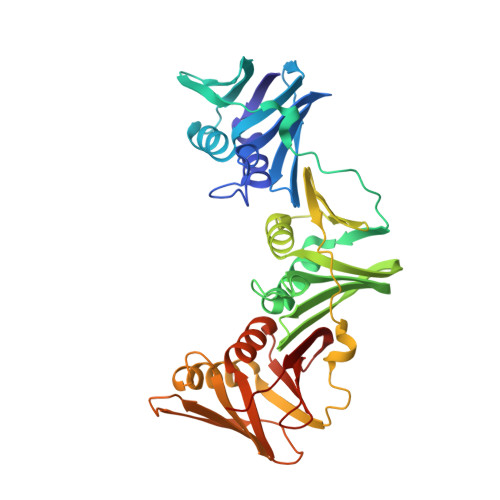
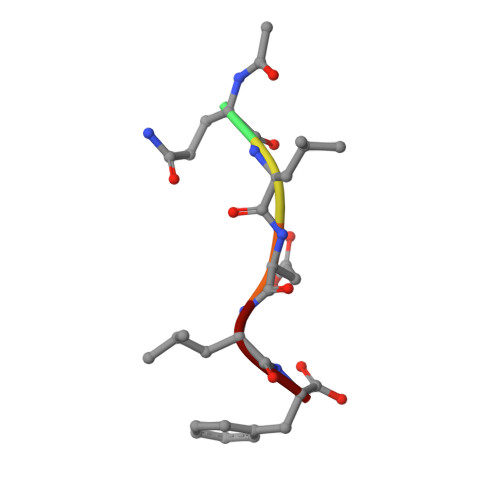
Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
Wolff, P., Olieric, V., Briand, J.P., Chaloin, O., Dejaegere, A., Dumas, P., Ennifar, E., Guichard, G., Wagner, J., Burnouf, D.Y.(2011) J Med Chem 54: 4627-4637
- PubMed: 21619076
- DOI: https://doi.org/10.1021/jm200311m
- Primary Citation of Related Structures:
3Q4J, 3Q4K, 3Q4L - PubMed Abstract:
The multimeric DNA sliding clamps confer high processivity to replicative DNA polymerases and are also binding platforms for various enzymes involved in DNA metabolism. These enzymes interact with the clamp through a small peptide that binds into a hydrophobic pocket which is a potential target for the development of new antibacterial compounds. Starting from a generic heptapeptide, we used a structure-based strategy to improve the design of new peptide ligands. Chemical modifications at specific residues result in a dramatic increase of the interaction as measured by SPR and ITC. The affinity of our best hits was improved by 2 orders of magnitude as compared to the natural ligand, reaching 10(-8) M range. The molecular basis of the interactions was analyzed by solving the co-crystal structures of the most relevant peptides bound to the clamp and reveals how chemical modifications establish new contacts and contributes to an increased affinity of the ligand.
- Architecture et Réactivité de l'ARN, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, Strasbourg, France.
Organizational Affiliation: