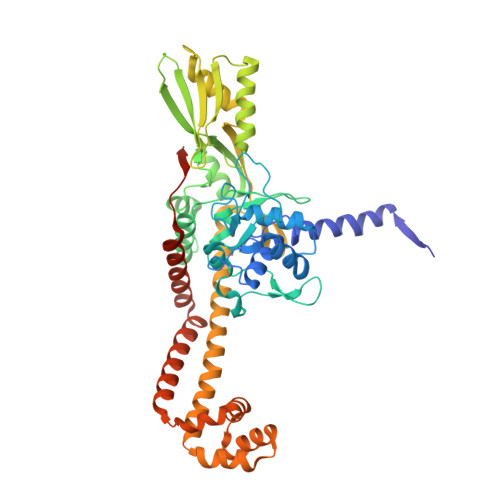
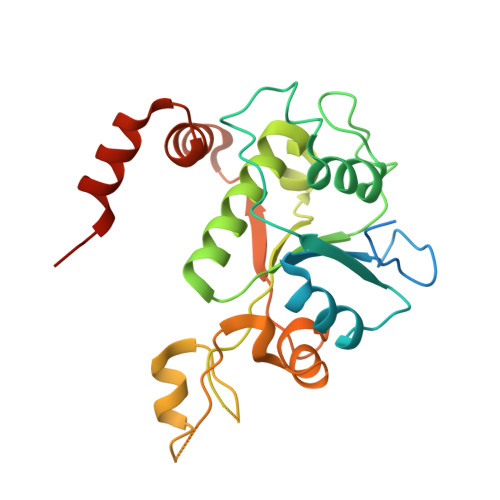
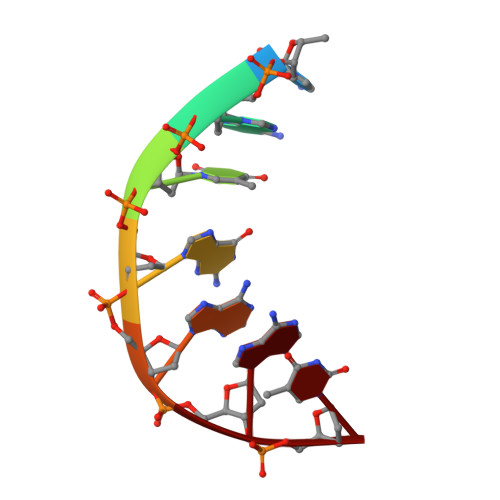
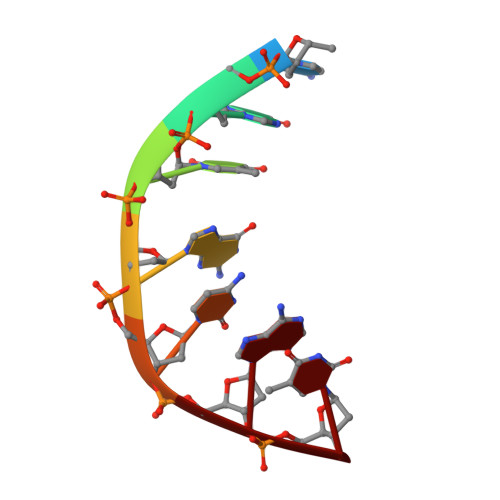
Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Laponogov, I., Pan, X.S., Veselkov, D.A., Cirz, R.T., Wagman, A., Moser, H.E., Fisher, L.M., Sanderson, M.R.(2016) Open Biol 6
- PubMed: 27655731
- DOI: https://doi.org/10.1098/rsob.160157
- Primary Citation of Related Structures:
3RAD, 4KPE, 4KPF - PubMed Abstract:
As part of a programme of synthesizing and investigating the biological properties of new fluoroquinolone antibacterials and their targeting of topoisomerase IV from Streptococcus pneumoniae, we have solved the X-ray structure of the complexes of two new 7,8-bridged fluoroquinolones (with restricted C7 group rotation favouring tight binding) in complex with the topoisomerase IV from S. pneumoniae and an 18-base-pair DNA binding site-the E-site-found by our DNA mapping studies to bind drug strongly in the presence of topoisomerase IV (Leo et al. 2005 J. Biol. Chem. 280, 14 252-14 263, doi:10.1074/jbc.M500156200). Although the degree of antibiotic resistance towards fluoroquinolones is much lower than that of β-lactams and a range of ribosome-bound antibiotics, there is a pressing need to increase the diversity of members of this successful clinically used class of drugs. The quinolone moiety of the new 7,8-bridged agents ACHN-245 and ACHN-454 binds similarly to that of clinafloxocin, levofloxacin, moxifloxacin and trovofloxacin but the cyclic scaffold offers the possibility of chemical modification to produce interactions with other topoisomerase residues at the active site.
Organizational Affiliation:
Randall Division of Cell and Molecular Biophysics, King's College, Guy's Campus, London Bridge, London SE1 1UL, UK Molecular and Clinical Sciences Research Institute, St George's, University of London, Cranmer Terrace, London SW17 0RE, UK.