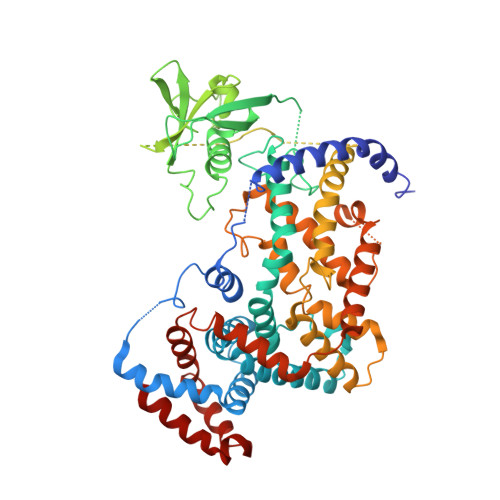
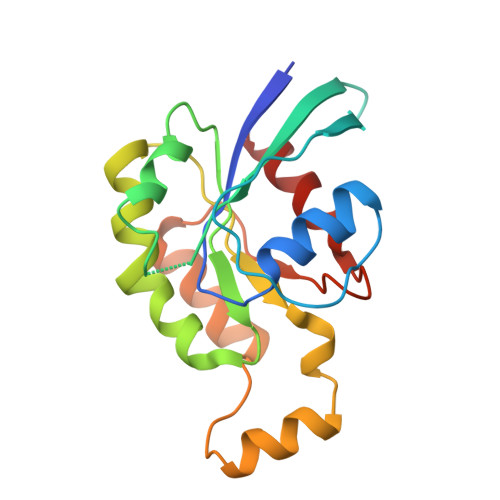
Plexins Are GTPase-Activating Proteins for Rap and Are Activated by Induced Dimerization.
Wang, Y., He, H., Srivastava, N., Vikarunnessa, S., Chen, Y.B., Jiang, J., Cowan, C.W., Zhang, X.(2012) Sci Signal 5: ra6-ra6
- PubMed: 22253263
- DOI: https://doi.org/10.1126/scisignal.2002636
- Primary Citation of Related Structures:
3RYT - PubMed Abstract:
Plexins are cell surface receptors that bind to semaphorins and transduce signals that regulate neuronal development, immune responses, and other processes. Signaling through plexins has been proposed to rely on specific guanosine triphosphatase (GTPase)-activating protein (GAP) activity for R-Ras and M-Ras. Activation of this GAP activity of plexins appears to require simultaneous binding of semaphorin to the plexin extracellular domain and of the Rho GTPases Rac1 or Rnd1 to the cytoplasmic region. However, GAP activity of plexins has eluded detection in several recent studies. We show that the purified cytoplasmic region of plexin uses a noncanonical catalytic mechanism to act as a GAP for Rap, but not for R-Ras or M-Ras. The RapGAP activity of plexins was autoinhibited and was activated by induced dimerization. Biochemical and crystallographic analyses demonstrated that binding of Rho GTPases did not directly contribute to activation of plexin RapGAP activity. Semaphorin stimulated the RapGAP activity of full-length plexin in cells, which was required for plexin-mediated neuronal growth cone collapse. Together, these findings define a pathway for plexin signaling and provide insights into the mechanism for semaphorin-induced activation of plexins.
Organizational Affiliation:
Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX 75063, USA.