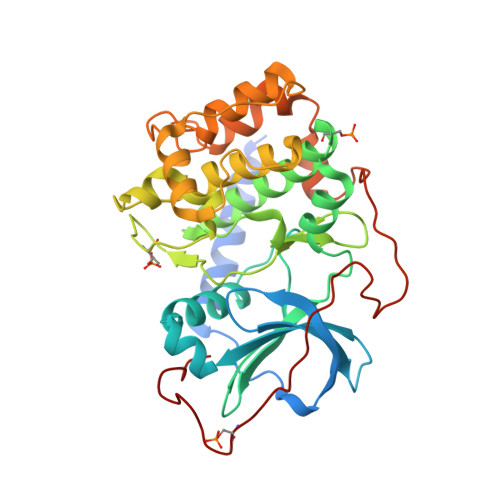
Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Gerlits, O., Das, A., Keshwani, M.M., Taylor, S., Waltman, M.J., Langan, P., Heller, W.T., Kovalevsky, A.(2014) Biochemistry 53: 3179-3186
- PubMed: 24786636
- DOI: https://doi.org/10.1021/bi5000965
- Primary Citation of Related Structures:
4IB0, 4IB1, 4IB3, 4O21, 4O22 - PubMed Abstract:
X-ray structures of several ternary product complexes of the catalytic subunit of cAMP-dependent protein kinase (PKAc) have been determined with no bound metal ions and with Na(+) or K(+) coordinated at two metal-binding sites. The metal-free PKAc and the enzyme with alkali metals were able to facilitate the phosphoryl transfer reaction. In all studied complexes, the ATP and the substrate peptide (SP20) were modified into the products ADP and the phosphorylated peptide. The products of the phosphotransfer reaction were also found when ATP-γS, a nonhydrolyzable ATP analogue, reacted with SP20 in the PKAc active site containing no metals. Single turnover enzyme kinetics measurements utilizing (32)P-labeled ATP confirmed the phosphotransferase activity of the enzyme in the absence of metal ions and in the presence of alkali metals. In addition, the structure of the apo-PKAc binary complex with SP20 suggests that the sequence of binding events may become ordered in a metal-free environment, with SP20 binding first to prime the enzyme for subsequent ATP binding. Comparison of these structures reveals conformational and hydrogen bonding changes that might be important for the mechanism of catalysis.
Organizational Affiliation:
Biology and Soft Matter Division, Oak Ridge National Laboratory , Oak Ridge, Tennessee 37831, United States.