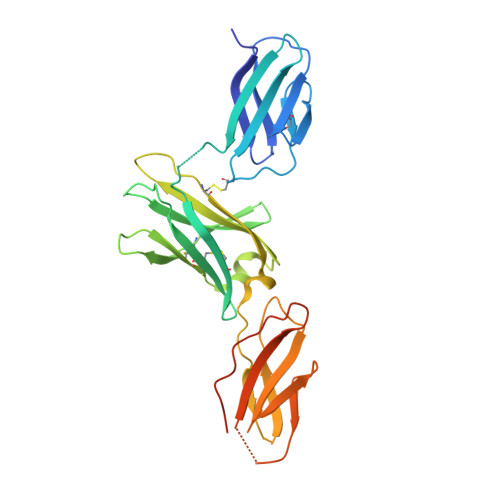
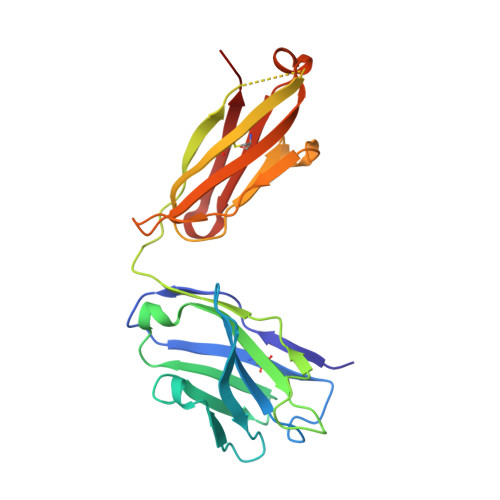
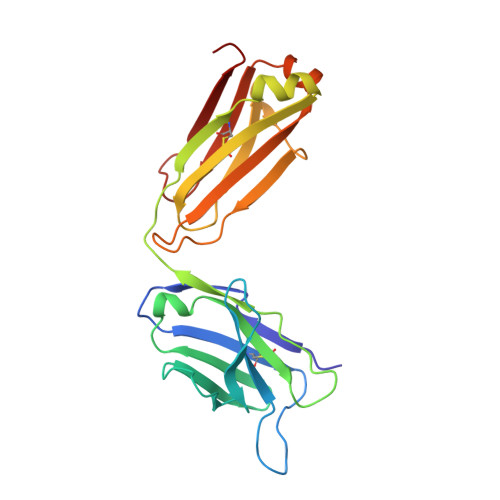
Dual mechanism of interleukin-3 receptor blockade by an anti-cancer antibody
Broughton, S.E., Hercus, T.R., Hardy, M.P., McClure, B.J., Nero, T.L., Dottore, M., Huynh, H., Braley, H., Barry, E.F., Kan, W.L., Dhagat, U., Scotney, P., Hartman, D., Busfield, S.J., Owczarek, C.M., Nash, A.D., Wilson, N.J., Parker, M.W., Lopez, A.F.(2014) Cell Rep 8: 410-419
- PubMed: 25043189
- DOI: https://doi.org/10.1016/j.celrep.2014.06.038
- Primary Citation of Related Structures:
4JZJ - PubMed Abstract:
Interleukin-3 (IL-3) is an activated T cell product that bridges innate and adaptive immunity and contributes to several immunopathologies. Here, we report the crystal structure of the IL-3 receptor α chain (IL3Rα) in complex with the anti-leukemia antibody CSL362 that reveals the N-terminal domain (NTD), a domain also present in the granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-5, and IL-13 receptors, adopting unique "open" and classical "closed" conformations. Although extensive mutational analyses of the NTD epitope of CSL362 show minor overlap with the IL-3 binding site, CSL362 only inhibits IL-3 binding to the closed conformation, indicating alternative mechanisms for blocking IL-3 signaling. Significantly, whereas "open-like" IL3Rα mutants can simultaneously bind IL-3 and CSL362, CSL362 still prevents the assembly of a higher-order IL-3 receptor-signaling complex. The discovery of open forms of cytokine receptors provides the framework for development of potent antibodies that can achieve a "double hit" cytokine receptor blockade.
- Australian Cancer Research Foundation Rational Drug Discovery Centre, St. Vincent's Institute of Medical Research, Fitzroy, VIC 3065, Australia.
Organizational Affiliation: