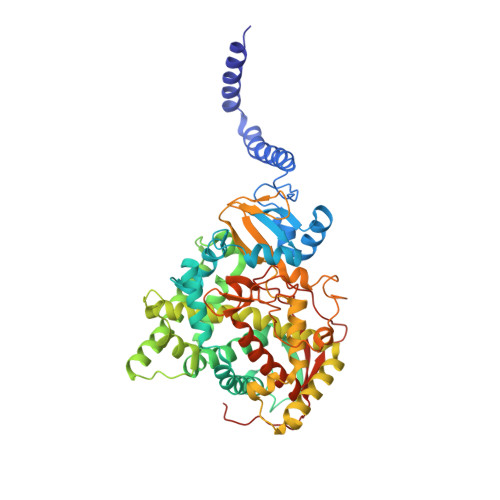
Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer.
Monk, B.C., Tomasiak, T.M., Keniya, M.V., Huschmann, F.U., Tyndall, J.D., O'Connell, J.D., Cannon, R.D., McDonald, J.G., Rodriguez, A., Finer-Moore, J.S., Stroud, R.M.(2014) Proc Natl Acad Sci U S A 111: 3865-3870
- PubMed: 24613931
- DOI: https://doi.org/10.1073/pnas.1324245111
- Primary Citation of Related Structures:
4LXJ, 5EQB - PubMed Abstract:
Bitopic integral membrane proteins with a single transmembrane helix play diverse roles in catalysis, cell signaling, and morphogenesis. Complete monospanning protein structures are needed to show how interaction between the transmembrane helix and catalytic domain might influence association with the membrane and function. We report crystal structures of full-length Saccharomyces cerevisiae lanosterol 14α-demethylase, a membrane monospanning cytochrome P450 of the CYP51 family that catalyzes the first postcyclization step in ergosterol biosynthesis and is inhibited by triazole drugs. The structures reveal a well-ordered N-terminal amphipathic helix preceding a putative transmembrane helix that would constrain the catalytic domain orientation to lie partly in the lipid bilayer. The structures locate the substrate lanosterol, identify putative substrate and product channels, and reveal constrained interactions with triazole antifungal drugs that are important for drug design and understanding drug resistance.
- Sir John Walsh Research Institute, Department of Oral Sciences, Faculty of Dentistry, and School of Pharmacy, University of Otago, Dunedin 9054, New Zealand.
Organizational Affiliation: