UD1
Query on UD1
Download Ideal Coordinates CCD File
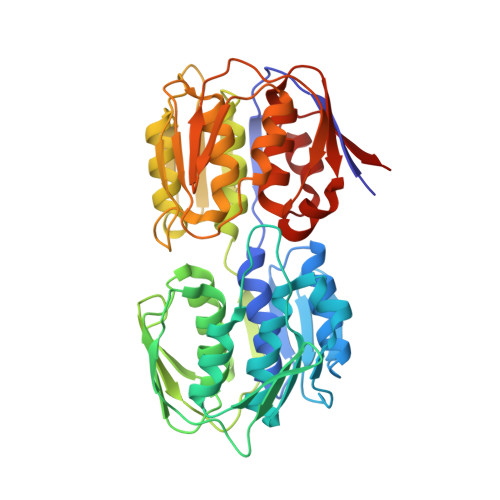
| G [auth A],
I [auth B],
L [auth C],
P [auth D] | URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE
C17 H27 N3 O17 P2
LFTYTUAZOPRMMI-CFRASDGPSA-N |  | |
PG4
Query on PG4
Download Ideal Coordinates CCD File
| E [auth A] | TETRAETHYLENE GLYCOL
C8 H18 O5
UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | |
FFQ
Query on FFQ
Download Ideal Coordinates CCD File
| F [auth A],
J [auth B],
M [auth C],
Q [auth D] | [(1R)-1-hydroxypropyl]phosphonic acid
C3 H9 O4 P
MVIJUJBSAAUHEM-GSVOUGTGSA-N |  | |
NA
Query on NA
Download Ideal Coordinates CCD File
| H [auth A],
K [auth B],
N [auth C],
O [auth C],
R [auth D] | SODIUM ION
Na
FKNQFGJONOIPTF-UHFFFAOYSA-N |  | |