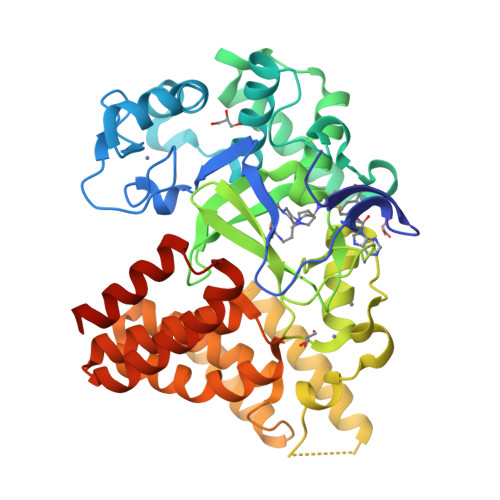
LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
Nguyen, H., Allali-Hassani, A., Antonysamy, S., Chang, S., Chen, L.H., Curtis, C., Emtage, S., Fan, L., Gheyi, T., Li, F., Liu, S., Martin, J.R., Mendel, D., Olsen, J.B., Pelletier, L., Shatseva, T., Wu, S., Zhang, F.F., Arrowsmith, C.H., Brown, P.J., Campbell, R.M., Garcia, B.A., Barsyte-Lovejoy, D., Mader, M., Vedadi, M.(2015) J Biol Chem 290: 13641-13653
- PubMed: 25825497
- DOI: https://doi.org/10.1074/jbc.M114.626861
- Primary Citation of Related Structures:
4WUY - PubMed Abstract:
SMYD2 is a lysine methyltransferase that catalyzes the monomethylation of several protein substrates including p53. SMYD2 is overexpressed in a significant percentage of esophageal squamous primary carcinomas, and that overexpression correlates with poor patient survival. However, the mechanism(s) by which SMYD2 promotes oncogenesis is not understood. A small molecule probe for SMYD2 would allow for the pharmacological dissection of this biology. In this report, we disclose LLY-507, a cell-active, potent small molecule inhibitor of SMYD2. LLY-507 is >100-fold selective for SMYD2 over a broad range of methyltransferase and non-methyltransferase targets. A 1.63-Å resolution crystal structure of SMYD2 in complex with LLY-507 shows the inhibitor binding in the substrate peptide binding pocket. LLY-507 is active in cells as measured by reduction of SMYD2-induced monomethylation of p53 Lys(370) at submicromolar concentrations. We used LLY-507 to further test other potential roles of SMYD2. Mass spectrometry-based proteomics showed that cellular global histone methylation levels were not significantly affected by SMYD2 inhibition with LLY-507, and subcellular fractionation studies indicate that SMYD2 is primarily cytoplasmic, suggesting that SMYD2 targets a very small subset of histones at specific chromatin loci and/or non-histone substrates. Breast and liver cancers were identified through in silico data mining as tumor types that display amplification and/or overexpression of SMYD2. LLY-507 inhibited the proliferation of several esophageal, liver, and breast cancer cell lines in a dose-dependent manner. These findings suggest that LLY-507 serves as a valuable chemical probe to aid in the dissection of SMYD2 function in cancer and other biological processes.
Organizational Affiliation:
From the Departments of Oncology Discovery, Structural Biology, Tailored Therapeutics, and Discovery Chemistry Research and Technologies, Eli Lilly and Company, Indianapolis, Indiana 46285, nguyen_anh-quan_hannah@lilly.com.