Structure of DrMEC-17 residue 2-185 at 2.85 Angstroms resolution
Xue, T.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Alpha-tubulin N-acetyltransferase 1 | 184 | Danio rerio | Mutation(s): 0 Gene Names: atat1, mec17 EC: 2.3.1.108 | 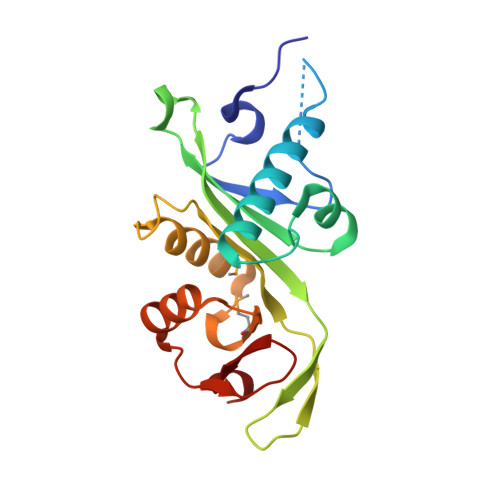 | |
UniProt | |||||
Find proteins for Q6PH17 (Danio rerio) Explore Q6PH17 Go to UniProtKB: Q6PH17 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6PH17 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ACO Query on ACO | C [auth A], E [auth B] | ACETYL COENZYME *A C23 H38 N7 O17 P3 S ZSLZBFCDCINBPY-ZSJPKINUSA-N |  | ||
| EPE Query on EPE | D [auth A], F [auth B] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 74.374 | α = 90 |
| b = 66.299 | β = 103.84 |
| c = 75.841 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data processing |
| HKL-2000 | data scaling |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Tianjin Natural Science Foundation | China | grant 14JCQNJC09300 |