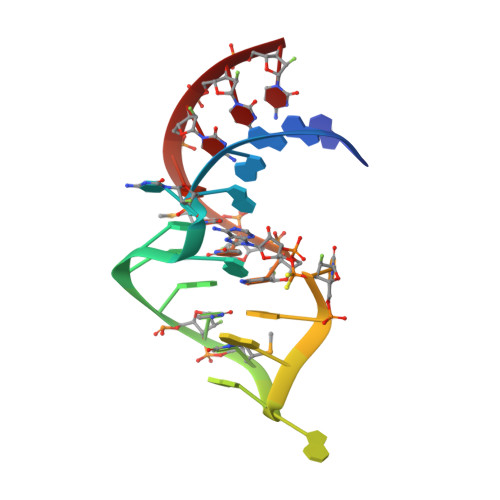
Evoking picomolar binding in RNA by a single phosphorodithioate linkage.
Abeydeera, N.D., Egli, M., Cox, N., Mercier, K., Conde, J.N., Pallan, P.S., Mizurini, D.M., Sierant, M., Hibti, F.E., Hassell, T., Wang, T., Liu, F.W., Liu, H.M., Martinez, C., Sood, A.K., Lybrand, T.P., Frydman, C., Monteiro, R.Q., Gomer, R.H., Nawrot, B., Yang, X.(2016) Nucleic Acids Res 44: 8052-8064
- PubMed: 27566147
- DOI: https://doi.org/10.1093/nar/gkw725
- Primary Citation of Related Structures:
5DO4 - PubMed Abstract:
RNA aptamers are synthetic oligonucleotide-based affinity molecules that utilize unique three-dimensional structures for their affinity and specificity to a target such as a protein. They hold the promise of numerous advantages over biologically produced antibodies; however, the binding affinity and specificity of RNA aptamers are often insufficient for successful implementation in diagnostic assays or as therapeutic agents. Strong binding affinity is important to improve the downstream applications. We report here the use of the phosphorodithioate (PS2) substitution on a single nucleotide of RNA aptamers to dramatically improve target binding affinity by ∼1000-fold (from nanomolar to picomolar). An X-ray co-crystal structure of the α-thrombin:PS2-aptamer complex reveals a localized induced-fit rearrangement of the PS2-containing nucleotide which leads to enhanced target interaction. High-level quantum mechanical calculations for model systems that mimic the PS2 moiety and phenylalanine demonstrate that an edge-on interaction between sulfur and the aromatic ring is quite favorable, and also confirm that the sulfur analogs are much more polarizable than the corresponding phosphates. This favorable interaction involving the sulfur atom is likely even more significant in the full aptamer-protein complexes than in the model systems.
Organizational Affiliation:
AM Biotechnologies, LLC, 12521 Gulf Freeway, Houston, TX 77034, USA.